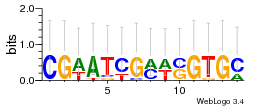
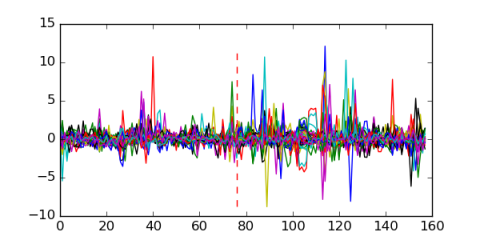
Module 255 Residual: 0.50
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 0.0000000015 | 0.0000000071 | 76 | 33 | 20160204 |
| MSMEG_3649 Uncharacterized protein | MSMEG_3020 Uncharacterized protein |
| MSMEG_3468 AMP-binding enzyme (AMP-dependent synthetase and ligase) | MSMEG_0422 PEP phosphonomutase (EC 2.7.8.23) (Transferase) |
| MSMEG_3565 Uncharacterized protein | MSMEG_0657 Rieske 2Fe-2S domain protein |
| MSMEG_3463 Putative esterase | MSMEG_5707 Cupin domain protein |
| MSMEG_6076 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (EC 2.7.7.60) (4-diphosphocytidyl-2C-methyl-D-erythritol synthase) (MEP cytidylyltransferase) | MSMEG_0427 Nitrite reductase (NAD(P)H) large subunit (Nitrite reductase [NAD(P)H], large subunit) (EC 1.7.1.4) |
| MSMEG_2213 O-methyltransferase | MSMEG_0428 Nitrite reductase (NAD(P)H) small subunit (Nitrite reductase [NAD(P)H] small subunit) (EC 1.7.1.4) |
| MSMEG_3466 Uncharacterized protein | MSMEG_4163 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100) (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_5975 Methyltransferase type 12, putative (Uncharacterized protein) | MSMEG_6551 Uncharacterized protein |
| MSMEG_6034 Uncharacterized protein | MSMEG_4164 3-hydroxybutyryl-CoA dehydratase (Enoyl-CoA hydratase/isomerase) (EC 4.2.1.17) |
| MSMEG_5977 dTDP-4-dehydrorhamnose 3,5-epimerase (EC 5.1.3.13) | MSMEG_0484 Formamidase (EC 3.5.1.49) |
| MSMEG_1007 Transport protein (Uncharacterized protein) | MSMEG_1092 Urease accessory protein UreF (Urease accessory protein UreF 1) |
| MSMEG_6071 Conserved hypothetical Zn-dependent hydrolase (Metallo-beta-lactamase superfamily protein) | MSMEG_1093 Urease (EC 3.5.1.5) |
| MSMEG_6073 Uncharacterized tRNA/rRNA methyltransferase MSMEG_6073/MSMEI_5913 (EC 2.1.1.-) | MSMEG_1091 Urease accessory protein UreE 1 (Urease accessory protein ureE 1) |
| MSMEG_6075 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (EC 4.6.1.12) | MSMEG_1096 Urease accessory protein UreD |
| MSMEG_6074 Cysteine--tRNA ligase (EC 6.1.1.16) (Cysteinyl-tRNA synthetase) (CysRS) | MSMEG_1094 Urease subunit alpha (EC 3.5.1.5) (Urea amidohydrolase subunit alpha) |
| MSMEG_2025 Putative IclR family protein transcriptional regulator (Transcriptional regulator IclR) | MSMEG_1095 Urease accessory protein UreG |
| MSMEG_0420 Uncharacterized protein |
KO_stationary.phase_rep1
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
M..smegmatis.Acid.NO.treated
Mycobacterium.smegmatis.Spheroplast.induced
X3dbfcy5plogcy3slide603norm
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
MB100pruR.vs..MB100..rep.3
Wild.type.rep1.x
Biological.replicate.4.of.6..kanamycin.treatment
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
X3dbfcy5plogcy3slide601norm
Wild.type.rep1.y
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.3.of.6..kanamycin.treatment
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep1
Biological.replicate.6.of.7..falcarinol.treatment
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.4
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
dcpA.knockout.rep2
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..kanamycin.treatment
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
X4XR.rep1
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Wild.type.rep3
WT.vs..DamtR.N.surplus_dyeswap_rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.6..negative.control
Biological.replicate.7.of.7..panaxydol.treatment
Plogcy5plogcy3slide113norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments