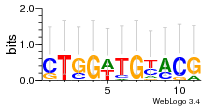
Module 273 Residual: 0.49
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.49 | 8.4e-16 | 2 | 82 | 35 | 20160204 |
| MSMEG_0784 Acetate kinase (EC 2.7.2.1) (Acetokinase) | MSMEG_1226 Sulfatase-modifying factor 1 |
| MSMEG_1495 HAD-superfamily protein hydrolase subfamily protein IA, variant 3, putative | MSMEG_3670 Small multidrug resistance protein (Transporter, small multidrug resistance (SMR) family protein) |
| MSMEG_0783 Phosphate acetyltransferase (Putative phosphate acetyltransferase Pta (Phosphotransacetylase)) (EC 2.3.1.8) | MSMEG_1928 Protein serine/threonine phosphatase (EC 3.1.3.16) (Regulatory protein) |
| MSMEG_5844 Uncharacterized protein | MSMEG_5330 Glutamine-fructose-6-phosphate transaminase (Isomerizing) (EC 2.6.1.16) (SIS domain protein) |
| MSMEG_0921 Uncharacterized protein | MSMEG_5358 Acetamidase/Formamidase family protein |
| MSMEG_3504 Probable membrane protein | MSMEG_5230 Uncharacterized protein |
| MSMEG_0922 Deoxyribose-phosphate aldolase (DERA) (EC 4.1.2.4) (2-deoxy-D-ribose 5-phosphate aldolase) (Phosphodeoxyriboaldolase) | MSMEG_6576 Pyridoxamine 5'-phosphate oxidase family protein (Pyridoxamine 5'-phosphate oxidase-related FMN-binding protein) |
| MSMEG_5847 Adenylosuccinate lyase (ASL) (EC 4.3.2.2) (Adenylosuccinase) | MSMEG_6142 UDP-glucose 4-epimerase (EC 5.1.3.2) (UDP-galactose 4-epimerase) (Uridine diphosphate galactose 4-epimerase) |
| MSMEG_5083 Uncharacterized protein | MSMEG_2267 Tetratricopeptide repeat domain protein |
| MSMEG_2899 Oxidoreductase, short chain dehydrogenase/reductase family protein (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.-) | MSMEG_3850 Alkanesulfonate monooxygenase (EC 1.14.14.5) (Alkanesulfonate monooxygenase family protein) |
| MSMEG_2270 Uncharacterized protein | MSMEG_2268 Rieske (2Fe-2S) region (Uncharacterized protein) |
| MSMEG_5229 Uncharacterized protein | MSMEG_2269 NHL repeat protein |
| MSMEG_4972 Acetyltransferase | MSMEG_2037 Uncharacterized protein |
| MSMEG_6338 Phosphoglycerate mutase (EC 5.4.2.-) (Phosphoglycerate mutase family protein, putative) | MSMEG_5846 Cytochrome P450 126 cyp126 (EC 1.14.13.151) (NikQ protein) |
| MSMEG_6337 Acyl-CoA dehydrogenase FadE36 (Phosphotransferase enzyme family protein) | MSMEG_4878 2Fe-2S iron-sulfur cluster binding domain protein (Ferredoxin) |
| MSMEG_3505 6-aminohexanoate-cyclic-dimer hydrolase (EC 3.5.2.12) (Amidase) (EC 3.5.1.4) | MSMEG_3608 Acetyl-CoA acetyltransferase |
| MSMEG_1695 Phosphoglucomutase/phosphomannomutase (Phosphoglucomutase/phosphomannomutase alpha/beta/alpha domain I) (EC 5.4.2.8) | MSMEG_6820 5'-nucleotidase surE (EC 3.1.3.5) (Acid phosphatase SurE) (EC 3.1.3.2) |
| MSMEG_1225 Uncharacterized protein |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.without.N_rep2
Biological.replicate.3.of.7..ethambutol.treatment
Mycobacterium.smegmatis.Spheroplast.induced
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
KO_log.phase_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
Wild.type.rep3
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Wild.type.rep2.y
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
Slow.vs..Fast.Replicate.2
X2.5..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..ethambutol.treatment
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X3dbfcy5plogcy3slide603norm
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
X4XR.rep2
dosR.WT_Replicate2
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
dcpA.knockout.rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.5.of.6..kanamycin.treatment
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments