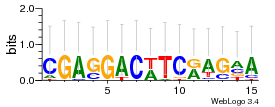
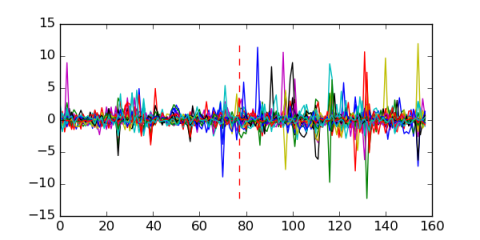
Module 329 Residual: 0.48
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.48 | 2.9e-19 | 0.00000000017 | 77 | 32 | 20160204 |
| MSMEG_3704 3-alpha-hydroxysteroid dehydrogenase | MSMEG_0874 Transcriptional regulator, GntR family protein |
| MSMEG_3705 MFS-type efflux pump MSMEG_3705 | MSMEG_0873 Uncharacterized protein |
| MSMEG_6846 Putative C4 decarboxylate transport protein (Tripartite ATP-independent periplasmic transporter DctQ component) | MSMEG_4610 IgiC, putative |
| MSMEG_3513 Enhanced intracellular survival protein (Enhanced intracellular survival protein Eis) | MSMEG_5562 Ribonuclease H (RNase H) (EC 3.1.26.4) |
| MSMEG_4609 Transcriptional regulator LysR family protein | MSMEG_3345 Transcriptional regulator, GntR family protein |
| MSMEG_3729 Catalase-peroxidase 3 (CP 3) (EC 1.11.1.21) (Peroxidase/catalase 3) | MSMEG_3346 Putative hydroxyacid aldolase |
| MSMEG_4504 Chaperone protein DnaJ | MSMEG_5195 Uncharacterized protein |
| MSMEG_3427 Beta-lactamase-like protein (Metal dependent hydrolase) | MSMEG_6430 Uncharacterized protein |
| MSMEG_3426 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) | MSMEG_1837 Secreted protein |
| MSMEG_5812 Carveol dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.275) | MSMEG_0777 F420-dependent glucose-6-phosphate dehydrogenase (FGD) (G6PD) (EC 1.1.98.2) |
| MSMEG_5813 Carveol dehydrogenase (EC 1.1.1.275) (Short-chain dehydrogenase/reductase SDR) | MSMEG_3486 Catalase (EC 1.11.1.6) (Catalase KatA) |
| MSMEG_0218 3-demethylubiquinone-9 3-methyltransferase | MSMEG_3485 RNA polymerase sigma factor |
| MSMEG_0217 Alcohol dehydrogenase B (EC 1.1.1.1) (Zinc-type alcohol dehydrogenase AdhD) | MSMEG_2581 Acetyltransferase |
| MSMEG_6214 GatB/Yqey (GatB/Yqey domain protein) | MSMEG_3765 TetR-family transcriptional regulator (Transcriptional regulator) |
| MSMEG_3947 6-phosphofructokinase isozyme 2 (EC 2.7.1.11) | MSMEG_3762 ABC transporter ATP-binding protein |
| MSMEG_4502 Ribosomal RNA small subunit methyltransferase E (EC 2.1.1.193) | MSMEG_3763 Transport permease protein |
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X4XR.rep1
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate3
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
KO_log.phase_rep2
KO_log.phase_rep1
Wild.type.rep1.y
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
stphcy5plogcy3slide487norm
Biological.replicate.2.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DamtR.N.surplus_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.2
Plogcy5plogcy3slide113norm
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
Biological.replicate.2.of.6..isoniazid.treatment
WT_log.phase_rep2
WT.with.vs..without.N_dye.swap_rep1
X3dbfcy3plogcy5slide607norm
Biological.replicate.4.of.7..panaxydol.treatment
X4dbfcy5plogcy3slide480norm
Biological.replicate.2.of.7..ethambutol.treatment
X2.5..vs..50..air.saturation.Replicate.3
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Biological.replicate.3.of.7..falcarinol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.6..kanamycin.treatment
Wild.type.rep3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
X2XR.rep1
X2XR.rep2
WT.vs..DglnR.N_starvation_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Comments
Log in to post comments