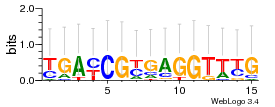
Module 339 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 7.5e-17 | 0.000000067 | 80 | 22 | 20160204 |
| MSMEG_6505 Nitroreductase NfnB (NR NfnB) (EC 1.-.-.-) (FMN-dependent NAD(P)H nitroreductase) | MSMEG_6365 Bacterial proteasome activator |
| MSMEG_6655 Uncharacterized protein | MSMEG_2012 Histidinol dehydrogenase (HDH) (EC 1.1.1.23) |
| MSMEG_6503 Transcriptional regulator, TetR family (Transcriptional regulator, TetR family protein) | MSMEG_2011 Transcriptional regulator LacI family (EC 5.1.1.1) (Transcriptional regulator, LacI family protein) |
| MSMEG_1512 dTDP-glucose 4,6-dehydratase (EC 4.2.1.46) | MSMEG_6366 O-antigen export system, ATP-binding protein (O-antigen/lipopolysaccharide ABC transporter ATP-binding protein RfbE) (EC 3.6.3.40) |
| MSMEG_1510 dTDP-4-dehydrorhamnose 3,5-epimerase (EC 5.1.3.13) (Thymidine diphospho-4-keto-rhamnose 3,5-epimerase) (dTDP-4-keto-6-deoxyglucose 3,5-epimerase) (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) (dTDP-L-rhamnose synthase) | MSMEG_6361 Transcriptional regulator, MarR family (Transcriptional regulator, MarR family protein) |
| MSMEG_1342 UPF0336 protein MSMEG_1342 | MSMEG_6362 Quinone oxidoreductase (EC 1.6.5.5) |
| MSMEG_1329 Major facilitator superfamily MFS_1 (Major facilitator superfamily protein MFS_1, putative) | MSMEG_6044 Periplasmic binding proteins and sugar binding domain of the LacI family protein, putative |
| MSMEG_1340 UPF0336 protein MSMEG_1340 | MSMEG_6369 O-antigen export system, permease protein (O-antigen/lipopolysaccharide ABC transporter permease protein RfbD) |
| MSMEG_1341 MaoC family protein (MaoC-like dehydratase) | MSMEG_0167 Transmembrane transport protein |
| MSMEG_0491 Transcriptional regulator, LacI family (Transcriptional regulator, LacI family protein) | MSMEG_1330 MarR-family protein transcriptional regulator |
| MSMEG_6367 Galactofuranosyl transferase GlfT1 (GalTr) (EC 2.4.1.287) (Beta-D-(1->5)galactofuranosyltransferase) (Beta-D-(1->6)galactofuranosyltransferase) (Rhamnopyranosyl-N-acetylglucosaminyl-diphospho-decaprenol beta-1,3/1,4-galactofuranosyltransferase) (UDP-Galf:alpha-3-L-rhamnosyl-alpha-D-GlcNAc-pyrophosphate polyprenol, UDP-galactofuranosyl transferase) | MSMEG_1339 50S ribosomal protein L33 1 |
Wild.type.rep1.y
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Wild.type.rep2.y
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Biological.replicate.4.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.30min
X2XR.rep1
dosR.WT_Replicate2
dosR.WT_Replicate1
Biological.replicate.2.of.6..kanamycin.treatment
Biological.replicate.2.of.7..panaxydol.treatment
dosR.WT_Replicate4
X3dbfcy5plogcy3slide601norm
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
Biological.replicate.1.of.6..negative.control
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DamtR.N.surplus_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.7.of.7..panaxydol.treatment
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.5.of.7..panaxydol.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_dyeswap_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
Wild.type.rep1.x
X2XR.rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.3.of.6..negative.control
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments