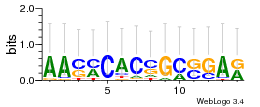
Module 347 Residual: 0.51
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.51 | 0.00005 | 0.00035 | 79 | 27 | 20160204 |
| MSMEG_2956 Fatty-acid-CoA ligase FadD9 (EC 6.2.1.3) (NAD dependent epimerase/dehydratase family protein) | MSMEG_4762 ABC transporter binding protein |
| MSMEG_2839 Transcriptional accessory protein | MSMEG_4761 ABC transporter ATP-binding protein (Spermidine/putrescine ABC transporter ATPase subunit) (EC 3.6.3.31) |
| MSMEG_4688 DSBA oxidoreductase (Uncharacterized protein) | MSMEG_5310 SAM-dependent methyltransferase |
| MSMEG_4689 Uncharacterized protein | MSMEG_0542 Antar domain protein |
| MSMEG_4179 Conserved integral membrane protein (Probable conserved integral membrane protein) | MSMEG_0541 Uncharacterized protein |
| MSMEG_5955 Protein-tyrosine kinase, putative (Uncharacterized protein) | MSMEG_1655 Alpha/beta hydrolase fold protein (Hydrolase, alpha/beta fold family protein, putative) |
| MSMEG_5954 Cell surface polysaccharide biosynthesis, putative | MSMEG_1656 Exodeoxyribonuclease III (EC 3.1.11.2) (Exodeoxyribonuclease III Xth) |
| MSMEG_0194 Cutinase Cut1 (EC 3.1.1.74) (Serine esterase, cutinase family protein) | MSMEG_1657 Tryptophan--tRNA ligase (EC 6.1.1.2) (Tryptophanyl-tRNA synthetase) |
| MSMEG_0195 Steroid monooxygenase | MSMEG_6242 Alcohol dehydrogenase, iron-containing (EC 1.1.1.1) (Iron-containing alcohol dehydrogenase) (EC 1.1.1.202) |
| MSMEG_1102 Amidohydrolase 3 (Twin-arginine translocation pathway signal) | MSMEG_6240 Uncharacterized protein |
| MSMEG_3090 ABC-type sugar transporter, permease protein (EC 3.6.3.17) (Ribose transport system permease protein RbsC) | MSMEG_6241 ATPase associated with various cellular activities AAA_5 (ATPase associated with various cellular activities, AAA-5) |
| MSMEG_3091 Ribose transport ATP-binding protein RbsA (EC 3.6.3.17) | MSMEG_1658 Conserved integral membrane protein (Ribonuclease, putative) |
| MSMEG_4764 ABC transporter permease protein | MSMEG_6586 Alpha/beta fold hydrolase (EC 3.-.-.-) (Alpha/beta hydrolase, putative) |
| MSMEG_4763 ABC transporter permease protein |
Bedaquiline.vs..DMSO.30min
WT.vs..DamtR.without.N_rep1
KO_log.phase_rep1
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate2
KO_log.phase_rep2
Wild.type.rep2.y
dosR.WT_Replicate4
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
Plogcy5plogcy3slide113norm
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.4.of.6..negative.control
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
WT_log.phase_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Biological.replicate.2.of.7..ethambutol.treatment
WT_log.phase_rep1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT.vs..DamtR.N.surplus_rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.N.surplus_dyeswap_rep2
X2XR.rep1
X2XR.rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.5.of.6..kanamycin.treatment
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..falcarinol.treatment
Comments
Log in to post comments