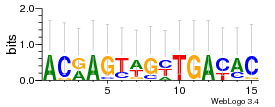
Module 363 Residual: 0.48
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.48 | 0.000000000034 | 0.00000000049 | 83 | 21 | 20160204 |
| MSMEG_0561 Putative TetR family protein receptor protein | MSMEG_0295 Oxidoreductase, FAD-binding |
| MSMEG_0560 Dihydrodipicolinate reductase (Dihydrodipicolinate reductase, N-terminus domain protein) (EC 1.17.1.8) | MSMEG_2313 |
| MSMEG_0292 Uncharacterized protein | MSMEG_0293 Rieske (2Fe-2S) region (EC 1.14.12.12) (Rieske [2Fe-2S] domain protein) |
| MSMEG_1032 Uncharacterized protein | MSMEG_5173 O-methyltransferase Omt (O-methyltransferase, putative) |
| MSMEG_0300 Amidohydrolase 2 (Amidohydrolase family protein) | MSMEG_5343 Uncharacterized protein |
| MSMEG_2400 50S ribosomal protein L28 | MSMEG_5172 Uncharacterized protein |
| MSMEG_1691 Transcriptional regulatory protein | MSMEG_5449 Uncharacterized protein |
| MSMEG_1692 ECF-family protein RNA polymerase sigma factor (Putative ECF-family RNA polymerase sigma factor) (EC 2.7.7.6) | MSMEG_2402 Dak phosphatase (Dihydroxyacetone kinase) |
| MSMEG_0294 3-oxoacyl-(Acyl-carrier-protein) reductase (EC 1.1.1.100) (Short-chain dehydrogenase/reductase SDR) | MSMEG_0299 Rieske (2Fe-2S) region (EC 1.14.12.12) (Rieske [2Fe-2S] domain protein) |
| MSMEG_0297 Amidohydrolase | MSMEG_0298 Uncharacterized protein |
| MSMEG_1690 RNA polymerase sigma factor |
KO_stationary.phase_rep1
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.without.N_rep2
dosR.WT_Replicate4
Biological.replicate.4.of.6..isoniazid.treatment
MB100pruR.vs..MB100..rep.3
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
X3dbfcy5plogcy3slide601norm
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
Bedaquiline.vs..DMSO.15min
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
Sub.MIC.INH
X0.6..vs..50..air.saturation.Replicate.5
Biological.replicate.6.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Plogcy5plogcy3slide113norm
Wild.type.rep2.y
Biological.replicate.5.of.6..kanamycin.treatment
stphcy3plogcy5slide581norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
Wild.type.rep2.x
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
X2XR.rep1
X2XR.rep2
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments