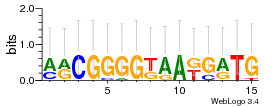
Module 369 Residual: 0.53
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.53 | 5.4e-31 | 0.0018 | 82 | 28 | 20160204 |
| MSMEG_0947 Acyltransferase (Phospholipid/glycerol acyltransferase) (EC 2.-.-.-) | MSMEG_5053 Short chain alcohol dehydrogenase (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_0946 NAD dependent epimerase/dehydratase family protein | MSMEG_3676 Phenoxybenzoate dioxygenase beta subunit (EC 1.-.-.-) (Putative Iron-sulfur oxidoreductase) (EC 1.14.12.7) |
| MSMEG_3675 Metal-dependent hydrolase (Uncharacterized protein) | MSMEG_3671 Integral membrane protein |
| MSMEG_2310 | MSMEG_3673 4-alpha-glucanotransferase (EC 2.4.1.25) |
| MSMEG_4734 Uncharacterized protein | MSMEG_1585 Cupin 2, conserved barrel (Cupin domain protein) |
| MSMEG_1624 Universal stress protein family protein (UspA domain protein) | MSMEG_5546 Conserved membrane protein (Membrane spanning protein) |
| MSMEG_5256 (2Z,6E)-farnesyl diphosphate synthase (EC 2.5.1.68) (Short-chain Z-isoprenyl diphosphate synthase) (Z-FPP synthase) (Z-FPPS) (Z-Polyprenyl diphosphate synthase) (Z-isoprenyl diphosphate synthase) | MSMEG_0814 Membrane protein |
| MSMEG_4733 Conserved hypothetical transmembrane protein (Putative membrane protein) | MSMEG_0817 LysR-family protein transcriptional regulator (LysR-family transcriptional regulator) |
| MSMEG_1574 Glutamate decarboxylase (EC 4.1.1.15) | MSMEG_0816 FAD-dependent pyridine nucleotide-disulfide oxidoreductase (EC 1.14.13.22) (Flavin-binding monooxygenase) |
| MSMEG_5548 Precorrin-6A synthase (Deacetylating) (Precorrin-6A synthase deacetylating) (EC 2.1.1.152) | MSMEG_5582 Uncharacterized protein |
| MSMEG_5251 Uncharacterized protein | MSMEG_5257 Channel protein, hemolysin III family (Channel protein, hemolysin III family protein) |
| MSMEG_1300 Uncharacterized protein | MSMEG_4280 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.-) |
| MSMEG_5012 Trans-sialidase, putative (Uncharacterized protein) | MSMEG_5549 Uncharacterized protein |
| MSMEG_5010 Uncharacterized protein | MSMEG_1030 Cyclohexanone monooxygenase (EC 1.14.13.22) (Monooxygenase) |
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.4.of.6..negative.control
Plogcy5plogcy3slide110norm
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate3
dosR.WT_Replicate2
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
rel.knockout.rep2
Biological.replicate.3.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep2
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.6.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
dosR.WT_Replicate4
Plogcy5plogcy3slide113norm
Biological.replicate.1.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
Sub.MIC.INH
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_dyeswap_rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X2XR.rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
Biological.replicate.2.of.6..negative.control
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments