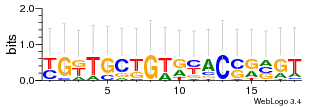
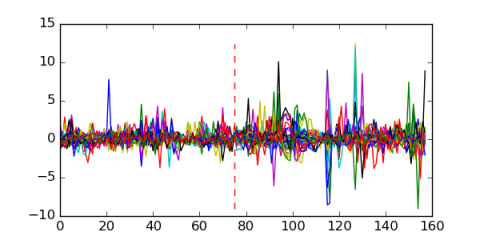
Module 384 Residual: 0.47
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.47 | 1200 | 13000 | 75 | 31 | 20160204 |
| MSMEG_3647 Glycogen accumulation regulator GarA | MSMEG_6628 Transcriptional regulator, TetR family (Transcriptional regulator, TetR family protein) |
| MSMEG_3646 Transcriptional regulator, MerR family (Transcriptional regulator, MerR family protein) | MSMEG_5884 3-hydroxyisobutyrate dehydrogenase family protein (6-phosphogluconate dehydrogenase NAD-binding protein) (EC 1.1.1.31) |
| MSMEG_3682 Lipolytic enzyme, G-D-S-L (Uncharacterized protein) | MSMEG_4026 Uncharacterized protein |
| MSMEG_6885 Coenzyme F420-dependent N5 N10-methylene tetrahydromethanopterin reductase-like protein (MmcI protein) | MSMEG_0810 Putative pyrimidine permease RutG (Uracil-xanthine permease) |
| MSMEG_6754 MaoC like domain protein (MaoC-like dehydratase) | MSMEG_1947 Glutaredoxin-like protein (Uncharacterized protein) |
| MSMEG_4400 Alcohol dehydrogenase, zinc-containing | MSMEG_3252 UPF0060 membrane protein MSMEG_3252/MSMEI_3169 |
| MSMEG_1063 Uncharacterized protein | MSMEG_5707 Cupin domain protein |
| MSMEG_6540 MCE-family protein MCE1a (Virulence factor Mce family protein) | MSMEG_6511 Acyl-CoA dehydrogenase (EC 1.3.8.-) (Acyl-CoA dehydrogenase domain protein) |
| MSMEG_5400 Dehydrogenase (FAD dependent oxidoreductase) | MSMEG_5410 Radical SAM (Radical SAM domain protein) |
| MSMEG_4682 Na+/H+ antiporter | MSMEG_1067 Glyoxalase/bleomycin resistance protein/dioxygenase (Uncharacterized protein) |
| MSMEG_2064 Response regulator receiver protein (Two-component system response regulator) | MSMEG_0564 Xanthine/uracil permease (Xanthine/uracil/vitamin C permease) |
| MSMEG_5875 Uncharacterized protein | MSMEG_6180 Secreted protein |
| MSMEG_3197 Lipase (Triacylglycerol lipase) (EC 3.1.1.3) | MSMEG_1318 Polysaccharide deacetylase (EC 3.5.2.16) (Polysaccharide deacetylase family protein) |
| MSMEG_5399 ATP-dependent DNA helicase RecQ (EC 3.6.1.-) | MSMEG_1505 Integral membrane protein (Putative integral membrane protein (DMT superfamily)) |
| MSMEG_4189 L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase (L-Cys:GlcN-Ins ligase) (EC 6.3.1.13) (Mycothiol ligase) (MSH ligase) | MSMEG_5064 Malyl-CoA lyase |
| MSMEG_6624 Uncharacterized protein |
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
KO_log.phase_rep1
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
Wild.type.rep2.x
Biological.replicate.5.of.6..isoniazid.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate2
dosR.WT_Replicate1
KO_log.phase_rep2
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.5
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Plogcy5plogcy3slide113norm
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.2
Biological.replicate.2.of.7..panaxydol.treatment
WT_stationary.phase_rep2
Biological.replicate.3.of.6..kanamycin.treatment
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.2.of.6..isoniazid.treatment
dosR.WT_Replicate3
Bedaquiline.vs..DMSO.60min
Biological.replicate.2.of.6..kanamycin.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X3dbfcy5plogcy3slide603norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
Wild.type.rep1.y
Biological.replicate.2.of.6..negative.control
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.5.of.6..kanamycin.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments