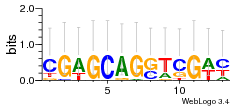
Module 401 Residual: 0.57
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.57 | 3.1e-21 | 1100 | 78 | 29 | 20160204 |
| MSMEG_6888 Integrase catalytic region (Transposase subunit) | MSMEG_2961 Protein translocase subunit SecD |
| MSMEG_3644 Transcriptional regulator, MerR family protein | MSMEG_5173 O-methyltransferase Omt (O-methyltransferase, putative) |
| MSMEG_3287 Alpha/beta hydrolase fold protein (EC 3.8.1.5) (Hydrolase, alpha/beta fold family protein) | MSMEG_2964 Adenine phosphoribosyltransferase (APRT) (EC 2.4.2.7) |
| MSMEG_3310 Integral membrane protein | MSMEG_1737 Pseudouridine synthase (RNA pseudouridine synthase family protein) |
| MSMEG_3288 LysM domain protein (Peptidoglycan-binding LysM) | MSMEG_6347 Uncharacterized protein |
| MSMEG_3648 Glycine cleavage system H protein | MSMEG_2786 Hydrolase, alpha/beta fold family protein |
| MSMEG_4296 Protease (Tripeptidyl-peptidase B Serine peptidase MEROPS family S33) | MSMEG_2785 Alpha-(1->3)-arabinofuranosyltransferase (EC 2.4.2.47) (Arabinofuranan 3-O-arabinosyltransferase) (arabinofuranosyltransferase C) (aftC) |
| MSMEG_3117 Cytochrome aa3 controlling protein (Unidentified antibiotic-transport integral membrane ABC transporter) | MSMEG_4965 Uncharacterized protein |
| MSMEG_0627 Glycosyl transferase, family protein 39 | MSMEG_0614 Putative S-adenosyl-L-methionine-dependent methyltransferase MSMEG_0614/MSMEI_0598 (EC 2.1.1.-) |
| MSMEG_2396 Putative conserved secreted protein (Uncharacterized protein) | MSMEG_6308 Uncharacterized protein |
| MSMEG_5417 Uncharacterized protein | MSMEG_2383 Glutamate--tRNA ligase (EC 6.1.1.17) (Glutamyl-tRNA synthetase) (GluRS) |
| MSMEG_1127 Probable conserved transmembrane protein (Putative conserved transmembrane protein) | MSMEG_2382 5-carboxymethyl-2-hydroxymuconate delta-isomerase (Putative 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase) |
| MSMEG_4089 Transposase subunit | MSMEG_2875 Alpha/beta hydrolase fold-3 (Esterase) |
| MSMEG_2963 Bacterial extracellular solute-binding protein, family protein 5 (Extracellular solute-binding protein) | MSMEG_5172 Uncharacterized protein |
| MSMEG_2962 Protein-export membrane protein SecF |
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
Biological.replicate.4.of.6..negative.control
X4XR.rep1
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.4.of.6..isoniazid.treatment
Wild.type.rep3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
KO_log.phase_rep2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
DglnR_without.vs..with.N_rep1
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.1.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
Biological.replicate.5.of.6..negative.control
X4dbfcy5plogcy3slide480norm
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
dosR.WT_Replicate2
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
Biological.replicate.2.of.6..negative.control
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.3.of.6..negative.control
Comments
Log in to post comments