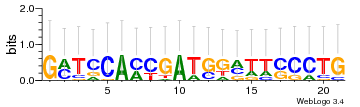
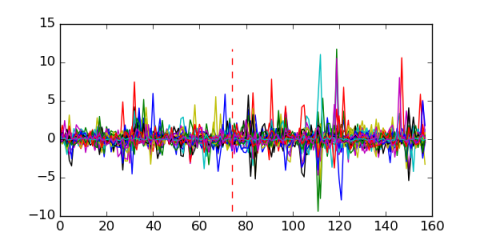
Module 412 Residual: 0.53
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.53 | 0.000000012 | 0.012 | 74 | 33 | 20160204 |
| MSMEG_2953 Ethyl tert-butyl ether degradation EthD | MSMEG_6491 Aldehyde dehydrogenase |
| MSMEG_5100 Pyruvate ferredoxin/flavodoxin oxidoreductase (EC 1.2.7.8) (Pyruvate ferredoxin/flavodoxin oxidoreductase family protein) | MSMEG_1322 ErfK/YbiS/YcfS/YnhG family protein |
| MSMEG_1591 | MSMEG_1734 Uncharacterized protein |
| MSMEG_1823 Uncharacterized protein | MSMEG_5258 Steroid Delta-isomerase (EC 5.3.3.1) (Steroid delta-isomerase) |
| MSMEG_2496 D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding protein (NAD-binding protein) | MSMEG_1310 TnpC protein |
| MSMEG_5447 Dolichyl-phosphate-mannose--protein O-mannosyl transferase (Dolichyl-phosphate-mannose-protein mannosyltransferase) | MSMEG_6134 NADP-dependent fatty aldehyde dehydrogenase (EC 1.2.1.4) |
| MSMEG_4719 Peptidase S9, prolyl oligopeptidase | MSMEG_0449 Drug resistance transporter EmrB/QacA subfamily (Transporter, major facilitator family protein, putative) |
| MSMEG_4392 Organophopsphate acid anhydrase | MSMEG_2495 Decarboxylase |
| MSMEG_6654 TnpC protein | MSMEG_1137 Amino acid permease-associated region |
| MSMEG_3228 Acyl-CoA thioesterase II (Acyl-CoA thioesterase II TesB1) | MSMEG_4720 Uncharacterized protein |
| MSMEG_2998 Uncharacterized protein | MSMEG_2642 Uncharacterized protein |
| MSMEG_2064 Response regulator receiver protein (Two-component system response regulator) | MSMEG_2497 Amidase (EC 3.5.1.4) (Amidase family protein) |
| MSMEG_6199 Transcriptional regulator WhiB | MSMEG_1724 Uncharacterized protein |
| MSMEG_2004 Arabinose-proton symporter | MSMEG_6339 Metallo-beta-lactamase family protein |
| MSMEG_0742 LysR-family transcriptional regulator (Probable transcriptional regulatory protein) | MSMEG_2441 Signal peptidase I (EC 3.4.21.89) |
| MSMEG_1455 Drug resistance transporter EmrB/QacA subfamily (Putative multidrug resistance protein) | MSMEG_4407 Uncharacterized protein |
| MSMEG_6492 GAF domain protein, putative |
Bedaquiline.vs..DMSO.30min
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
Biological.replicate.5.of.6..kanamycin.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
Biological.replicate.2.of.7..panaxydol.treatment
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
MB100pruR.vs..MB100.rep.2
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep3
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.3
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Slow.vs..Fast.Replicate.2
X2.5..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..ethambutol.treatment
X3dbfcy5plogcy3slide603norm
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
dosR.WT_Replicate2
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Comments
Log in to post comments