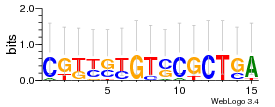
Module 415 Residual: 0.56
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.56 | 0.000041 | 0.038 | 80 | 26 | 20160204 |
| MSMEG_1935 TetR-family protein transcriptional regulator (TetR-family transcriptional regulator) | MSMEG_6832 Uncharacterized protein |
| MSMEG_1936 Uncharacterized protein | MSMEG_4631 Uncharacterized protein |
| MSMEG_6638 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase (EC 2.1.1.14) (Cobalamin-independent methionine synthase) (Methionine synthase, vitamin-B12 independent isozyme) | MSMEG_1075 1,4-dihydroxy-2-naphthoyl-CoA synthase (DHNA-CoA synthase) (EC 4.1.3.36) |
| MSMEG_6351 Putative phenylalanine aminotransferase (EC 2.6.1.-) | MSMEG_1076 Uncharacterized protein |
| MSMEG_6352 Uncharacterized protein | MSMEG_1810 Uncharacterized protein |
| MSMEG_6353 Delta3,5-delta2,4-dienoyl-CoA isomerase (EC 5.3.3.-) (Enoyl-CoA hydratase EchA21) (EC 4.2.1.17) | MSMEG_2116 PTS system, glucose-specific IIBC component (EC 2.7.1.-) (EC 2.7.1.69) |
| MSMEG_1644 Uncharacterized protein | MSMEG_2117 Beta-glucoside-specific EII permease (PTS system sugar phosphotransferase component IIA) (EC 2.7.1.69) |
| MSMEG_6053 Cob(II)yrinic acid a,c-diamide reductase (EC 1.16.8.1) | MSMEG_1754 Uncharacterized protein |
| MSMEG_6237 Uncharacterized protein | MSMEG_2118 Glucosamine-6-phosphate deaminase (EC 3.5.99.6) (GlcN6P deaminase) (GNPDA) (Glucosamine-6-phosphate isomerase) |
| MSMEG_1453 Citrate transporter (Putative integral membrane protein) | MSMEG_2119 N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25) (GlcNAc 6-P deacetylase) |
| MSMEG_1454 Uncharacterized protein | MSMEG_5024 T/U mismatch-specific DNA glycosylase |
| MSMEG_1781 Uncharacterized protein | MSMEG_0006 DNA gyrase subunit A (EC 5.99.1.3) |
| MSMEG_6833 Alcohol dehydrogenase (EC 1.1.1.1) | MSMEG_0005 DNA gyrase subunit B (EC 5.99.1.3) |
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.without.N_rep2
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
DglnR_without.vs..with.N_rep2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.3.of.6..kanamycin.treatment
X4XR.rep1
Biological.replicate.1.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.2
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
Sub.MIC.INH
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DamtR.N.surplus_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Biological.replicate.7.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
dcpA.knockout.rep2
dcpA.knockout.rep1
X4dbfcy5plogcy3slide480norm
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.4
WT_log.phase_rep1
DglnR_without.vs..with.N_rep1
X2.5..vs..50..air.saturation.Replicate.1
WT_stationary.phase_rep2
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
dosR.WT_Replicate3
X3dbfcy5plogcy3slide603norm
Biological.replicate.4.of.6..kanamycin.treatment
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.2.of.6..negative.control
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.1.of.6..kanamycin.treatment
dosR.WT_Replicate2
Comments
Log in to post comments