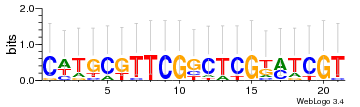
Module 432 Residual: 0.50
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 2e-24 | 0.0000022 | 78 | 34 | 20160204 |
| MSMEG_1883 Choline/carnitine/betaine transporter (Glycine betaine transporter OpuD) | MSMEG_0499 Amino acid permease-associated region |
| MSMEG_5461 Uncharacterized protein | MSMEG_4303 Methyltransferase (Methyltransferase type 11) (EC 2.1.1.80) |
| MSMEG_5460 Uncharacterized protein | MSMEG_6538 3-oxoacyl-(Acyl-carrier-protein) reductase, putative (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_1288 Uncharacterized protein | MSMEG_5601 Uncharacterized protein |
| MSMEG_3447 Putative two-component system response regulator, LuxR family (Two-component system response regulator) | MSMEG_0500 Regulator of polyketide synthase expression, putative |
| MSMEG_0823 Uncharacterized protein | MSMEG_1517 SPFH domain / Band 7 family protein (Spfh domain/band 7 family protein, putative) |
| MSMEG_4737 Uncharacterized protein | MSMEG_1412 Amino acid permease (Cationic amino acid transport integral membrane protein RocE) |
| MSMEG_1287 Cyclase/dehydrase superfamily protein | MSMEG_5801 Hydroxylase (Uncharacterized protein) |
| MSMEG_1286 Sulfatase (EC 3.1.6.6) (Sulfatase family protein) | MSMEG_5802 Oxidoreductase, short chain dehydrogenase/reductase family protein (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) |
| MSMEG_1740 Dehydrogenase/reductase SDR family protein member 4 (EC 1.1.1.184) (Short-chain dehydrogenase/reductase SDR) | MSMEG_6321 Glycerol dehydratase large subunit |
| MSMEG_6318 Putative diol dehydratase reactivation protein (Uncharacterized protein) | MSMEG_6320 Diol dehydrase gamma subunit (Propanediol dehydratase, small subunit) (EC 4.2.1.30) |
| MSMEG_1413 Ornithine--oxo-acid transaminase (EC 2.6.1.13) | MSMEG_1520 50S ribosomal protein L36 |
| MSMEG_1519 Translation initiation factor IF-1 | MSMEG_1135 Uncharacterized protein |
| MSMEG_0497 Glycerol dehydratase large subunit (Propanediol dehydratase medium subunit) (EC 4.2.1.30) | MSMEG_5026 Uncharacterized protein |
| MSMEG_0496 Coenzyme B12-dependent glycerol dehydrogenase small subunit (Propanediol dehydratase small subunit) (EC 4.2.1.30) | MSMEG_5285 Patatin (Phospholipase, patatin family protein) |
| MSMEG_4304 Regulatory protein | MSMEG_0483 Major facilitator superfamily MFS_1 (Shikimate transporter) |
| MSMEG_1738 Probable conserved transmembrane protein (Putative conserved transmembrane protein) | MSMEG_3662 LysM domain protein (Mannose-binding lectin) |
mc2155.vs.Dcrp1.replicate.1
WT.vs..DamtR.without.N_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Wild.type.rep2.x
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate2
KO_log.phase_rep2
Wild.type.rep2.y
Wild.type.rep1.y
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
X4XR.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.5.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DamtR.N.surplus_rep2
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
Slow.vs..Fast.Replicate.3
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
X3dbfcy5plogcy3slide603norm
dcpA.knockout.rep1
X4dbfcy5plogcy3slide480norm
X3dbfcy5plogcy3slide601norm
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Biological.replicate.2.of.7..ethambutol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
WT_stationary.phase_rep2
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep2
X2XR.rep1
X2XR.rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
Sub.MIC.INH
Plogcy5plogcy3slide113norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments