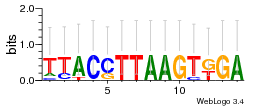
Module 440 Residual: 0.53
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.53 | 0.00000004 | 0.014 | 78 | 28 | 20160204 |
| MSMEG_2953 Ethyl tert-butyl ether degradation EthD | MSMEG_1104 Uncharacterized protein |
| MSMEG_3406 Putative transcriptional regulator, TetR family protein (TetR-family transcriptional regulator) | MSMEG_1785 Uncharacterized protein |
| MSMEG_3405 Methylenomycin A resistance protein | MSMEG_1782 Oxidoreductase, short chain dehydrogenase/reductase family protein (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) |
| MSMEG_6444 YiaAB two helix domain protein | MSMEG_5035 Uncharacterized protein |
| MSMEG_3150 3-oxoacyl-[acyl-carrier-protein] reductase FabG (EC 1.1.1.100) (3-ketoacyl-acyl carrier protein reductase) (Beta-Ketoacyl-acyl carrier protein reductase) (Beta-ketoacyl-ACP reductase) | MSMEG_1783 Uncharacterized protein |
| MSMEG_3152 Ferrochelatase (EC 4.99.1.1) (Heme synthase) (Protoheme ferro-lyase) | MSMEG_4438 Uncharacterized protein |
| MSMEG_3247 Branched-chain amino acid ABC transporter substrate-binding protein | MSMEG_3151 Enoyl-[acyl-carrier-protein] reductase [NADH] (EC 1.3.1.9) (NADH-dependent enoyl-ACP reductase) |
| MSMEG_5420 Tat-translocated enzyme | MSMEG_4437 Cytochrome c oxidase subunit 1 (EC 1.9.3.1) (Cytochrome c oxidase, subunit I) |
| MSMEG_3240 DNA-binding response regulator, LuxR family protein | MSMEG_3239 Putative signal transduction histidine kinase (Two-component system sensor kinase) |
| MSMEG_3249 Branched chain amino acid transport permease (Branched-chain amino acid ABC transporter, permease protein) | MSMEG_3250 ABC transporter, ATP-binding protein (Branched-chain amino acid ABC transporter (LivG)) |
| MSMEG_3248 ABC transporter branched chain amino acid transport permease (Inner-membrane translocator) | MSMEG_3251 Branched-chain amino acid ABC transporter ATP-binding protein |
| MSMEG_0094 Uncharacterized protein | MSMEG_3451 Short-chain dehydrogenase/reductase (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_0097 Oxidoreductase, zinc-binding dehydrogenase family protein (Putative quinone oxidoreductase (NADPH:quinone oxidoreductase) (Zeta-crystallin)) (EC 1.6.5.5) | MSMEG_3452 TetR family protein transcriptional regulator |
| MSMEG_0096 Peroxisomal hydratase-dehydrogenase-epimerase (EC 1.1.1.-) (EC 4.2.1.-) | MSMEG_2019 Short-chain dehydrogenase/reductase SDR (EC 1.1.1.100) |
Bedaquiline.vs..DMSO.30min
Wild.type.rep2.y
dcpA.knockout.rep2
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.2.of.6..kanamycin.treatment
KO_log.phase_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.x
Wild.type.rep1.y
DglnR_without.vs..with.N_rep2
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.4.of.6..kanamycin.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
X3dbfcy5plogcy3slide601norm
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.2.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
WT.with.vs..without.N_dye.swap_rep2
WT.vs..DamtR.N.surplus_dyeswap_rep2
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.3.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
X3dbfcy5plogcy3slide603norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
Biological.replicate.2.of.6..negative.control
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Sub.MIC.INH
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
dosR.WT_Replicate2
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
Comments
Log in to post comments