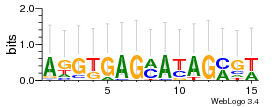
Module 474 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 4.6e-26 | 0.0000000065 | 75 | 30 | 20160204 |
| MSMEG_5483 Porin MspC | MSMEG_5403 Probable cobalt/nickel-exporting P-type ATPase (EC 3.6.3.-) (Cation-transporting P-type ATPase CtpD) |
| MSMEG_0944 DNA binding domain, excisionase family protein | MSMEG_3783 Acyl-CoA dehydrogenase (EC 1.3.99.-) |
| MSMEG_4175 Transcriptional regulator, ArsR family protein | MSMEG_5214 RNA polymerase sigma factor |
| MSMEG_1863 | MSMEG_4934 ATP:cob(I)alamin adenosyltransferase (EC 2.5.1.17) (EC 4.2.1.28) |
| MSMEG_1862 Transposase | MSMEG_2489 Transcriptional regulator, GntR family (Transcriptional regulator, GntR family protein, putative) |
| MSMEG_6674 Transcriptional regulator, LysR family (Transcriptional regulator, LysR family protein) | MSMEG_3475 Uncharacterized protein |
| MSMEG_4871 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.178) (EC 1.1.1.35) (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.-) | MSMEG_6328 Uncharacterized protein |
| MSMEG_2044 Integral membrane transport protein (Major facilitator superfamily MFS_1) | MSMEG_6361 Transcriptional regulator, MarR family (Transcriptional regulator, MarR family protein) |
| MSMEG_2045 Monoxygenase (Xenobiotic compound monooxygenase, DszA family) | MSMEG_3183 L-threonine dehydratase (EC 4.3.1.19) (Threonine deaminase) |
| MSMEG_5407 Integral membrane protein (Putative integral membrane protein) | MSMEG_3975 Putative regulatory protein (Regulatory protein, putative) |
| MSMEG_5406 Uncharacterized protein | MSMEG_2177 Fmnh2-utilizing oxygenase |
| MSMEG_5405 Transcriptional regulator, ArsR family protein (Transcriptional regulatory protein) | MSMEG_4869 LAO/AO transport system ATPase (EC 2.7.-.-) |
| MSMEG_6330 Prephenate dehydrogenase (EC 1.3.1.12) | MSMEG_1274 Gluconolactonase (EC 3.1.1.17) |
| MSMEG_2651 Alkanesulfonate monooxygenase family protein | MSMEG_5000 LysR-family transcriptional regulator (Transcriptional regulator, LysR family protein) |
| MSMEG_2650 Acyl-CoA dehydrogenase (Putative acyl-CoA dehydrogenase) | MSMEG_5003 O-methyltransferase family 3 (EC 2.1.1.104) (O-methyltransferase, family protein 3) |
Bedaquiline.vs..DMSO.30min
WT.vs..DamtR.without.N_rep1
KO_log.phase_rep1
WT.vs..DamtR.without.N_rep2
Plogcy5plogcy3slide110norm
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
Biological.replicate.2.of.6..kanamycin.treatment
Wild.type.rep2.y
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..panaxydol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
Biological.replicate.5.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.2
Biological.replicate.6.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.4
X0.6..vs..50..air.saturation.Replicate.4
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..negative.control
stphcy3plogcy5slide581norm
WT_log.phase_rep1
X3dbfcy5plogcy3slide603norm
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Biological.replicate.3.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
dosR.WT_Replicate4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Biological.replicate.4.of.6..kanamycin.treatment
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_dyeswap_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
X2XR.rep1
X2XR.rep2
WT.vs..DglnR.N_starvation_dye.swap_rep2
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments