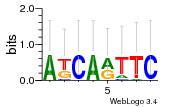
Module 490 Residual: 0.56
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.56 | 2.1e-22 | 8 | 78 | 31 | 20160204 |
| MSMEG_1883 Choline/carnitine/betaine transporter (Glycine betaine transporter OpuD) | MSMEG_4434 Cof protein (Cof-like hydrolase) |
| MSMEG_1915 Anti-sigma factor RshA (Regulator of SigH) (Sigma-H anti-sigma factor RshA) | MSMEG_6167 Bacterial type II secretion system protein F domain (Type II secretion system protein) |
| MSMEG_5243 Helix-turn-helix motif (Pyridoxamine 5'-phosphate oxidase-related, FMN-binding protein) | MSMEG_6624 Uncharacterized protein |
| MSMEG_5242 Diacylglycerol O-acyltransferase (EC 2.3.1.20) | MSMEG_2194 MerR-family protein transcriptional regulator (MerR-family transcriptional regulator) |
| MSMEG_1185 Transcriptional regulator, AsnC family protein | MSMEG_5742 Acyltransferase domain protein |
| MSMEG_0538 MarR family transcriptional regulator (Regulatory protein, MarR) | MSMEG_6904 Inositol-3-phosphate synthase (IPS) (EC 5.5.1.4) (Myo-inositol 1-phosphate synthase) (MI-1-P synthase) (MIP synthase) |
| MSMEG_4391 Acyl-CoA dehydrogenase (EC 1.3.8.-) (Acyl-CoA dehydrogenase-family protein) | MSMEG_4632 Saccharopine dehydrogenase (EC 1.5.1.7) |
| MSMEG_5956 NAD dependent epimerase/dehydratase family protein (UDP-glucose 4-epimerase, GalE5) | MSMEG_1741 TetR-family protein transcriptional regulator |
| MSMEG_6168 Bacterial type II secretion system protein F domain | MSMEG_0852 CBS domain protein |
| MSMEG_4138 Aliphatic nitrilase (EC 3.5.5.7) | MSMEG_4389 Coenzyme F420-dependent N5,N10-methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase (EC 1.14.14.5) (Monooxygenase, NtaA/SnaA/SoxA family protein) |
| MSMEG_1742 Oxidoreductase (Oxidoreductase FAD-binding region) | MSMEG_2669 Dienelactone hydrolase (EC 3.1.1.45) (Hydrolase) |
| MSMEG_1743 Fatty acid desaturase | MSMEG_0146 Mosc domain protein |
| MSMEG_2252 Flavin-type hydroxylase | MSMEG_3852 Aliphatic sulfonate binding protein (Aliphatic sulfonates family ABC transporter, periplasmic ligand-binding protein) |
| MSMEG_2102 ABC transporter related protein (Nitrate transport ATP-binding protein NrtD) | MSMEG_4095 Putative monooxygenase |
| MSMEG_0537 Conserved hypothetical membrane protein (Uncharacterized protein) | MSMEG_5743 Patatin |
| MSMEG_3598 ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein (EC 3.6.3.17) (Periplasmic sugar-binding proteins) |
KO_stationary.phase_rep2
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Bedaquiline.vs..DMSO.15min
Biological.replicate.5.of.6..isoniazid.treatment
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.30min
Biological.replicate.4.of.6..isoniazid.treatment
dosR.WT_Replicate1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
KO_log.phase_rep1
DglnR_without.vs..with.N_rep1
Wild.type.rep1.x
Wild.type.rep1.y
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep3
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep2
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.3
Biological.replicate.1.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
X4XR.rep2
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
X3dbfcy5plogcy3slide603norm
mc2155.vs.Dcrp1.replicate.1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DglnR.N_starvation_rep2
Wild.type.rep2.x
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments