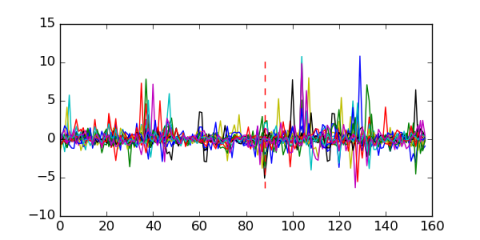
Module 492 Residual: 0.47
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.47 | 0.00000000000053 | 0.0000000000044 | 88 | 19 | 20160204 |
| MSMEG_5697 Conserved integral membrane protein (Integral membrane protein) | MSMEG_1682 Flavin-containing monooxygenase FMO |
| MSMEG_0660 Bacterial extracellular solute-binding protein | MSMEG_1681 Endoribonuclease L-PSP superfamily protein |
| MSMEG_6174 Transcriptional regulator, IclR family (Transcriptional regulator, IclR family protein) | MSMEG_0780 Phosphotransferase enzyme family protein |
| MSMEG_6175 2-keto-3-deoxy-galactonokinase (2-keto-3-deoxy-galactonokinase (2-dehydro-3-deoxygalactonokinase)) (EC 2.7.1.58) | MSMEG_6178 D-galactonate transporter (Major facilitator superfamily MFS_1) |
| MSMEG_6176 2-dehydro-3-deoxy-6-phosphogalactonate aldolase (EC 4.1.2.21) (KDPG and KHG aldolase) | MSMEG_0659 Polyamine ABC transporter permease (Polyamine ABC transporter permease protein) |
| MSMEG_6177 Galactonate dehydratase (EC 4.2.1.6) (Mandelate racemase/muconate lactonizing protein (Galactonate dehydratase)) | MSMEG_0658 Polyamine ABC-transporter, inner membrane subunit |
| MSMEG_1679 AmiB (Amidohydrolase AmiB1) | MSMEG_0662 Polyamine-transporting ATPase (EC 3.6.3.31) |
| MSMEG_5696 'Cold-shock' DNA-binding domain protein | MSMEG_0782 Aminotransferase class III (EC 2.6.1.-) (Aminotransferase class-III) |
| MSMEG_0661 Glutamate-1-semialdehyde 2,1-aminomutase (EC 5.4.3.8) | MSMEG_1677 Aspartate ammonia-lyase (EC 4.3.1.1) |
| MSMEG_0781 Amino acid permease (Amino acid permease-associated region) |
KO_stationary.phase_rep2
Biological.replicate.4.of.7..ethambutol.treatment
Wild.type.rep2.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep2
WT.vs..DamtR.N.surplus_rep2
stphcy5plogcy3slide487norm
X3dbfcy5plogcy3slide601norm
Biological.replicate.2.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.6.of.7..ethambutol.treatment
X4dbfcy5plogcy3slide901norm
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.without.N_rep1
WT.vs..DamtR.without.N_rep2
Biological.replicate.4.of.6..negative.control
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
Biological.replicate.1.of.7..panaxydol.treatment
Biological.replicate.3.of.6..negative.control
WT_log.phase_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
WT_log.phase_rep1
WT.with.vs..without.N_dye.swap_rep1
WT.with.vs..without.N_dye.swap_rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DglnR.N_starvation_rep2
X2XR.rep1
X2XR.rep2
Sub.MIC.INH
Mycobacterium.smegmatis.Spheroplast.induced
MB100pruR.vs..MB100..rep.3
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
DglnR_without.vs..with.N_rep1
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.2.of.6..isoniazid.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Wild.type.rep1.y
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep3
Biological.replicate.4.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.4
dcpA.knockout.rep2
dcpA.knockout.rep1
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.2
X4XR.rep2
X4XR.rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
WT.vs..DamtR.N.surplus_dyeswap_rep2
Biological.replicate.5.of.7..falcarinol.treatment
Biological.replicate.1.of.6..negative.control
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Comments
Log in to post comments