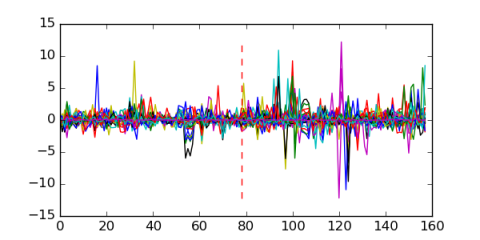
Module 508 Residual: 0.48
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.48 | 0.0000000000068 | 0.000000049 | 78 | 33 | 20160204 |
| MSMEG_3359 Cis-3-chloroacrylic acid dehalogenase, putative | MSMEG_2774 Diguanylate cyclase (Ggdef) domain protein |
| MSMEG_2956 Fatty-acid-CoA ligase FadD9 (EC 6.2.1.3) (NAD dependent epimerase/dehydratase family protein) | MSMEG_1266 ADP-ribosylation/Crystallin J1 (Ankyrin-repeat containing protein) |
| MSMEG_0866 DNA or RNA helicase of superfamily protein II | MSMEG_3616 Integral membrane protein (Transglycosylase-associated protein) |
| MSMEG_2184 Amino acid permease (Amino acid permease-associated region) | MSMEG_0155 Transcriptional regulator |
| MSMEG_2183 Uncharacterized protein | MSMEG_5881 Carbon monoxide dehydrogenase subunit G (Putative carbon monoxide dehydrogenase subunit G) |
| MSMEG_6311 Saccharopine dehydrogenase (EC 1.5.1.7) | MSMEG_5880 Nicotine dehydrogenase (Xanthine dehydrogenase, molybdenum binding subunit apoprotein) (EC 1.17.1.4) |
| MSMEG_3246 Response regulator (Two-component system response regulator) | MSMEG_5883 (2Fe-2S)-binding protein (EC 1.-.-.-) ([2Fe-2S] binding domain protein) |
| MSMEG_3455 Heat shock protein 15 | MSMEG_5882 Aerobic-type carbon monoxide dehydrogenase protein, FAD-binding subunit (Carbon monoxide dehydrogenase medium chain) (EC 1.2.99.2) |
| MSMEG_4954 Transcription termination factor Rho | MSMEG_4385 ABC transporter oligopeptide binding protein |
| MSMEG_4582 Uncharacterized protein (VWA containing CoxE-like protein) | MSMEG_4387 ABC transporter permease protein (Binding-protein-dependent transport systems inner membrane component) |
| MSMEG_5917 Uncharacterized protein | MSMEG_4386 ABC transporter permease protein (Oligopeptide ABC transporter, permease) (EC 3.6.3.24) |
| MSMEG_5879 D-alanyl-D-alanine dipeptidase (D-Ala-D-Ala dipeptidase) (EC 3.4.13.22) | MSMEG_1191 Uncharacterized protein |
| MSMEG_4583 ATPase associated with various cellular activities | MSMEG_1772 Uncharacterized protein |
| MSMEG_2798 Uncharacterized protein | MSMEG_1771 Methylase, putative |
| MSMEG_5934 Uncharacterized protein | MSMEG_1770 Uncharacterized protein |
| MSMEG_0156 Transcriptional regulator, LysR family protein | MSMEG_0167 Transmembrane transport protein |
| MSMEG_5936 Uncharacterized protein |
KO_stationary.phase_rep2
WT.vs..DglnR.N_starvation_rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.without.N_rep2
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
Biological.replicate.3.of.7..ethambutol.treatment
dosR.WT_Replicate2
KO_log.phase_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
rel.knockout.rep2
Wild.type.rep3
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
WT_log.phase_rep2
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.6.of.7..ethambutol.treatment
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Plogcy5plogcy3slide113norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
X0.6..vs..50..air.saturation.Replicate.4
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
WT.vs..DamtR.N.surplus_rep2
Comments
Log in to post comments