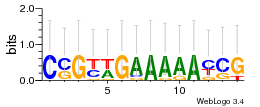
Module 544 Residual: 0.51
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.51 | 0.0000000000000012 | 0.00000067 | 81 | 26 | 20160204 |
| MSMEG_5104 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase (EC 2.3.1.117) (Tetrahydrodipicolinate N-succinyltransferase) (THDP succinyltransferase) (THP succinyltransferase) (Tetrahydropicolinate succinylase) | MSMEG_0743 Xanthine dehydrogenase |
| MSMEG_6297 Aldehyde Dehydrogenase (EC 1.2.-.-) (Aldehyde dehydrogenase) | MSMEG_1622 Putative DNA repair polymerase (Putative DNA-directed DNA polymerase) (EC 2.7.7.7) |
| MSMEG_5534 DNA helicase (EC 3.6.4.12) | MSMEG_5643 Uncharacterized protein |
| MSMEG_5536 Intracellular chorismate mutase (CM) (EC 5.4.99.5) | MSMEG_1619 Uncharacterized protein |
| MSMEG_2494 Peptidase, M24 family protein (EC 3.4.13.9) (Xaa-Pro aminopeptidase) | MSMEG_4040 Nitroreductase |
| MSMEG_2493 Aminotransferase, class I and II family protein (Transcriptional regulator, GntR family) (EC 2.6.1.39) | MSMEG_0998 Oxidoreductase, molybdopterin binding |
| MSMEG_2491 Acetylornithine deacetylase (Acetylornithine deacetylase/succinyl-diaminopimelate desuccinylase) (EC 3.5.1.-) | MSMEG_1757 ATP-dependent DNA helicase (DEAD/DEAH box helicase) |
| MSMEG_6296 5-exo-alcohol dehydrogenase (EC 1.1.1.-) (Alcohol dehydrogenase, zinc-containing) (EC 1.1.1.14) | MSMEG_1756 DNA glycosylase/endonuclease (EC 4.2.99.18) (Endonuclease VIII and dna n-glycosylase with an ap lyase activity) |
| MSMEG_6299 D-isomer specific 2-hydroxyacid dehydrogenase (EC 1.1.1.-) (Glyoxylate reductase) (EC 1.1.1.26) | MSMEG_1759 Uncharacterized protein |
| MSMEG_6298 Citrate lyase beta chain (EC 4.1.3.6) (Malyl-CoA lyase) | MSMEG_1795 2-deoxy-scyllo-inosamine dehydrogenase (EC 1.1.1.-) |
| MSMEG_1620 Uncharacterized protein | MSMEG_1796 Membrane protein (Uncharacterized protein) |
| MSMEG_0720 Integral membrane protein | MSMEG_4279 Adenylate and Guanylate cyclase catalytic domain protein (Uncharacterized protein) |
| MSMEG_3968 Uncharacterized protein | MSMEG_0049 Pirin domain protein |
WT.vs..DamtR.without.N_rep1
Biological.replicate.3.of.6..kanamycin.treatment
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
Mycobacterium.smegmatis.Spheroplast.induced
Plogcy5plogcy3slide110norm
Biological.replicate.4.of.7..ethambutol.treatment
Wild.type.rep3
X2XR.rep1
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
KO_log.phase_rep1
Wild.type.rep2.x
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X0.6..vs..50..air.saturation.Replicate.5
Slow.vs..Fast.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
X0.6..vs..50..air.saturation.Replicate.4
Biological.replicate.5.of.7..panaxydol.treatment
Plogcy5plogcy3slide111norm
WT_log.phase_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.5.of.7..falcarinol.treatment
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X2XR.rep2
Biological.replicate.1.of.7..panaxydol.treatment
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate3
dosR.WT_Replicate2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments