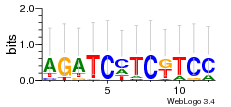
Module 545 Residual: 0.62
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.62 | 0.0000000049 | 0.000000037 | 81 | 33 | 20160204 |
| MSMEG_6529 Uncharacterized protein | MSMEG_0034 FHA domain-containing protein FhaB (FtsZ-interacting protein A) |
| MSMEG_6528 Uncharacterized protein | MSMEG_1223 Uncharacterized protein |
| MSMEG_6527 Transcriptional regulator, TetR family (Transcriptional regulator, TetR family protein) | MSMEG_4366 Serine/threonine-protein kinase PknD (EC 2.7.11.1) |
| MSMEG_6220 Lipoprotein (Putative conserved membrane protein) | MSMEG_3649 Uncharacterized protein |
| MSMEG_2494 Peptidase, M24 family protein (EC 3.4.13.9) (Xaa-Pro aminopeptidase) | MSMEG_0762 Cytochrome P450 FAS1 (EC 1.14.-.-) |
| MSMEG_2493 Aminotransferase, class I and II family protein (Transcriptional regulator, GntR family) (EC 2.6.1.39) | MSMEG_6530 Cytochrome C oxidase subunit III (EC 1.9.3.1) (Cytochrome c oxidase subunit III family protein) |
| MSMEG_2491 Acetylornithine deacetylase (Acetylornithine deacetylase/succinyl-diaminopimelate desuccinylase) (EC 3.5.1.-) | MSMEG_6453 Sulfate permease |
| MSMEG_6219 ATPase family protein associated with various cellular activities (AAA) | MSMEG_5258 Steroid Delta-isomerase (EC 5.3.3.1) (Steroid delta-isomerase) |
| MSMEG_6218 Secreted protein | MSMEG_4902 Beta-lactamase-like protein (EC 3.1.6.1) (Metal-dependent hydrolase of the beta-lactamase superfamily protein III) |
| MSMEG_0761 Conserved domain protein (Putative ferredoxin) | MSMEG_3910 Uncharacterized protein |
| MSMEG_6217 Conserved membrane protein (Integral membrane protein) | MSMEG_2156 NmrA-like protein (Uncharacterized protein) |
| MSMEG_6216 Integral membrane protein | MSMEG_2930 Uncharacterized protein |
| MSMEG_0033 Protein phosphatase 2C (Serine/threonine phosphatase PPP) (EC 3.1.3.16) | MSMEG_4755 Peptidase M20 |
| MSMEG_0032 Cell cycle protein, FtsW/RodA/SpoVE family protein (Cell division protein RodA) | MSMEG_0028 Serine/threonine-protein kinase PknB (EC 2.7.11.1) |
| MSMEG_0031 Penicillin-binding protein A (PBPA) | MSMEG_5168 Uncharacterized protein |
| MSMEG_0030 Serine/threonine protein kinase PknA (EC 2.7.1.-) (Serine/threonine-protein kinases pknA) (EC 2.7.11.1) | MSMEG_6827 Uncharacterized protein |
| MSMEG_0233 Lipoprotein Lpps |
KO_stationary.phase_rep1
Biological.replicate.5.of.7..falcarinol.treatment
KO_log.phase_rep1
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
dcpA.knockout.rep2
Biological.replicate.4.of.6..negative.control
Wild.type.rep2.x
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate3
dosR.WT_Replicate2
X4dbfcy5plogcy3slide480norm
KO_log.phase_rep2
Wild.type.rep2.y
dosR.WT_Replicate4
Wild.type.rep1.x
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Wild.type.rep3
MB100pruR.vs..MB100.rep.2
Biological.replicate.6.of.7..panaxydol.treatment
WT.vs..DamtR.without.N_rep2
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
X2XR.rep2
stphcy5plogcy3slide487norm
Biological.replicate.5.of.6..negative.control
Biological.replicate.5.of.6..kanamycin.treatment
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
X4XR.rep1
Sub.MIC.INH
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X3dbfcy5plogcy3slide601norm
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.1
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Biological.replicate.2.of.6..kanamycin.treatment
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
mc2155.vs.Dcrp1.replicate.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Bedaquiline.vs..DMSO.60min
Biological.replicate.4.of.7..falcarinol.treatment
Biological.replicate.5.of.6..isoniazid.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X3dbfcy5plogcy3slide603norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
X4dbfcy5plogcy3slide901norm
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.1.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_dyeswap_rep2
X2XR.rep1
Bedaquiline.vs..DMSO.15min
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
Biological.replicate.2.of.6..negative.control
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments