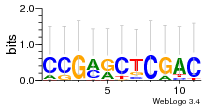
Module 547 Residual: 0.59
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.59 | 1.2e-16 | 630 | 79 | 33 | 20160204 |
| MSMEG_5481 SAF domain protein (Uncharacterized protein) | MSMEG_0171 Histone deacetylase superfamily protein |
| MSMEG_4901 Ribonuclease PH (RNase PH) (EC 2.7.7.56) (tRNA nucleotidyltransferase) | MSMEG_6851 PEBP family protein (Phosphatidylethanolamine-binding protein) |
| MSMEG_6766 ABC transporter related protein (ABC transporter, ATP-binding protein, NodI family protein) | MSMEG_4899 Non-canonical purine NTP pyrophosphatase (EC 3.6.1.19) (Non-standard purine NTP pyrophosphatase) (Nucleoside-triphosphate diphosphatase) (Nucleoside-triphosphate pyrophosphatase) |
| MSMEG_1490 3-oxoacyl-ACP synthase III | MSMEG_6872 Beta-lactamase (Uncharacterized protein) |
| MSMEG_1491 Histidinol-phosphate aminotransferase 2 (EC 2.6.1.9) | MSMEG_1489 Uncharacterized protein |
| MSMEG_4662 Deoxyribose-phosphate aldolase superfamily protein | MSMEG_6896 Single-stranded DNA-binding protein (SSB) |
| MSMEG_4663 Myo-inositol catabolism IolB protein (Protein IolB) | MSMEG_6895 30S ribosomal protein S18 2 |
| MSMEG_0922 Deoxyribose-phosphate aldolase (DERA) (EC 4.1.2.4) (2-deoxy-D-ribose 5-phosphate aldolase) (Phosphodeoxyriboaldolase) | MSMEG_6894 50S ribosomal protein L9 |
| MSMEG_4666 Inositol 2-dehydrogenase (EC 1.1.1.18) (Myo-inositol 2-dehydrogenase) (MI 2-dehydrogenase) | MSMEG_4002 Oxidoreductase, zinc-binding dehydrogenase family protein |
| MSMEG_4664 IolD protein (Thiamine pyrophosphate-requiring enzyme, putative acetolactate synthase) (EC 2.2.1.6) | MSMEG_4000 Hypothetical ABC transporter ATP-binding protein YphE |
| MSMEG_4665 IolE protein (Xylose isomerase domain protein TIM barrel) | MSMEG_4001 Permease protein of sugar ABC transporter (Ribose transport system permease protein RbsC) (EC 3.6.3.17) |
| MSMEG_5341 Alpha/beta superfamily hydrolase (Dipeptidyl aminopeptidase/acylaminoacyl peptidase) | MSMEG_0921 Uncharacterized protein |
| MSMEG_1992 L-carnitine dehydratase/bile acid-inducible protein F | MSMEG_0307 Transcriptional regulator, AraC family protein |
| MSMEG_4714 MoaC domain protein | MSMEG_4980 Major facilitator superfamily MFS_1 (Major facilitator superfamily protein MFS_1) |
| MSMEG_6644 Citrate (Pro-3S)-lyase (EC 4.1.3.-) (Malyl-CoA lyase) | MSMEG_5142 Beta-glucosidase (EC 3.2.1.21) |
| MSMEG_4857 Conserved domain protein (Ferredoxin) | MSMEG_6765 Transport permease protein |
| MSMEG_4856 Cytochrome P450 (Cytochrome P450 105Q4 Cyp105Q4) |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
M..smegmatis.Acid.NO.treated
Mycobacterium.smegmatis.Spheroplast.induced
Plogcy5plogcy3slide110norm
Biological.replicate.5.of.6..kanamycin.treatment
dosR.WT_Replicate2
dosR.WT_Replicate1
KO_log.phase_rep2
WT.vs..DglnR.N_starvation_dye.swap_rep1
Wild.type.rep2.x
Wild.type.rep1.x
DglnR_without.vs..with.N_rep1
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.7..ethambutol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.4.of.6..kanamycin.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
mc2155.vs.Dcrp1.replicate.2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.4.of.6..negative.control
mc2155.vs.Dcrp1.replicate.4
Plogcy5plogcy3slide111norm
Biological.replicate.3.of.6..kanamycin.treatment
X2.5..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.6.of.6..isoniazid.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.6.of.6..kanamycin.treatment
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.2.of.6..negative.control
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Sub.MIC.INH
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..falcarinol.treatment
Comments
Log in to post comments