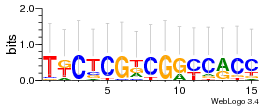
Module 549 Residual: 0.49
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.49 | 0.0027 | 6.4 | 76 | 31 | 20160204 |
| MSMEG_4770 Twin-arginine translocation pathway signal (Uncharacterized protein) | MSMEG_5793 Uncharacterized protein |
| MSMEG_6179 Acetyl-coenzyme A synthetase (AcCoA synthetase) (Acs) (EC 6.2.1.1) (Acetate--CoA ligase) (Acyl-activating enzyme) | MSMEG_5792 UPF0678 fatty acid-binding protein-like protein MSMEG_5792/MSMEI_5639 |
| MSMEG_6443 DNA polymerase IV (Pol IV) (EC 2.7.7.7) | MSMEG_1768 Multimeric flavodoxin WrbA (Uncharacterized protein) |
| MSMEG_4400 Alcohol dehydrogenase, zinc-containing | MSMEG_3820 Uncharacterized protein |
| MSMEG_4401 Phosphonoacetaldehyde hydrolase | MSMEG_1488 Acetolactate synthase |
| MSMEG_3115 Regulatory protein, DeoR | MSMEG_5817 Uncharacterized protein |
| MSMEG_0927 Uncharacterized protein | MSMEG_5451 Alkylated DNA repair protein |
| MSMEG_0926 Uncharacterized protein | MSMEG_6265 Transcriptional regulator (Transcriptional regulator, XRE family) |
| MSMEG_0929 ErfK/YbiS/YcfS/YnhG family protein (Lipoprotein lprQ) | MSMEG_1675 Pyrimidine-nucleoside phosphorylase (EC 2.4.2.2) (Thymidine phosphorylase) (EC 2.4.2.4) |
| MSMEG_0928 UDP-N-acetylenolpyruvoylglucosamine reductase (EC 1.3.1.98) (UDP-N-acetylmuramate dehydrogenase) | MSMEG_2746 Conserved hypothetical alanine and leucine rich protein |
| MSMEG_1433 Thioesterase | MSMEG_1134 Protease HtpX homolog (EC 3.4.24.-) |
| MSMEG_1765 Restriction endonuclease family protein | MSMEG_3482 Uncharacterized protein |
| MSMEG_1766 Uncharacterized protein | MSMEG_3481 o-succinylbenzoate synthase (EC 4.2.1.113) |
| MSMEG_1767 Uncharacterized protein | MSMEG_2587 Methionine aminopeptidase (MAP) (MetAP) (EC 3.4.11.18) (Peptidase M) |
| MSMEG_2376 Uncharacterized protein | MSMEG_2586 Uncharacterized protein |
| MSMEG_5791 Uncharacterized protein |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Biological.replicate.4.of.6..negative.control
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
MB100pruR.vs..MB100..rep.3
KO_log.phase_rep2
Biological.replicate.2.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_dyeswap_rep2
Wild.type.rep1.x
Wild.type.rep1.y
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.vs.Dcrp1.replicate.1
X3dbfcy3plogcy5slide607norm
dcpA.knockout.rep2
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.2.of.7..falcarinol.treatment
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
stphcy3plogcy5slide581norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
Biological.replicate.5.of.6..isoniazid.treatment
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep2
Slow.vs..Fast.Replicate.3
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
Wild.type.rep2.x
X2XR.rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Comments
Log in to post comments