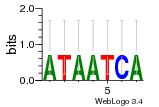
Module 607 Residual: 0.51
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.51 | 0.000000011 | 2800 | 87 | 18 | 20160204 |
| MSMEG_3894 Proteasome subunit alpha (EC 3.4.25.1) (20S proteasome alpha subunit) (Proteasome core protein PrcA) | MSMEG_3895 Proteasome subunit beta (EC 3.4.25.1) (20S proteasome beta subunit) (Proteasome core protein PrcB) |
| MSMEG_3859 Polyprenol monophosphomannose synthase (PPM synthase) (Polyprenol-P-Man synthase) (Ppm1) (EC 2.4.1.-) (Dolichol-phosphate mannose synthase) (EC 2.4.1.83) | MSMEG_3873 Cobalamin biosynthesis protein cobIJ (EC 2.1.1.130) (Precorrin-3B C17-methyltransferase) (EC 2.1.1.131) |
| MSMEG_3872 Precorrin-8X methylmutase (EC 5.4.1.2) | MSMEG_3861 Amidohydrolase 3 |
| MSMEG_3087 Preprotein translocase, SecG subunit (Protein-export membrane protein (Translocase subunit) SecG) | MSMEG_3860 Apolipoprotein N-acyltransferase (ALP N-acyltransferase) (EC 2.3.1.-) (LntMs) |
| MSMEG_3086 Triosephosphate isomerase (TIM) (EC 5.3.1.1) (Triose-phosphate isomerase) | MSMEG_1283 Antitoxin VapB |
| MSMEG_3148 Uncharacterized protein | MSMEG_3085 Phosphoglycerate kinase (EC 2.7.2.3) |
| MSMEG_3149 UPF0353 protein MSMEG_3149 | MSMEG_2134 Putative fluoride ion transporter CrcB |
| MSMEG_3147 ATPase, MoxR family protein | MSMEG_2135 Putative fluoride ion transporter CrcB |
| MSMEG_3084 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (EC 1.2.1.12) (NAD-dependent glyceraldehyde-3-phosphate dehydrogenase) | MSMEG_1284 Ribonuclease VapC (RNase VapC) (EC 3.1.-.-) (Endoribonuclease VapC) (Toxin VapC) |
Biological.replicate.4.of.7..ethambutol.treatment
Wild.type.rep2.y
Wild.type.rep2.x
MB100pruR.vs..MB100.rep.2
MB100pruR.vs..MB100.rep.1
Bedaquiline.vs..DMSO..45min
stphcy5plogcy3slide487norm
stphcy5plogcy3slide580norm
Biological.replicate.4.of.7..falcarinol.treatment
Biological.replicate.6.of.7..ethambutol.treatment
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.without.N_rep1
WT.vs..DamtR.without.N_rep2
Biological.replicate.4.of.6..negative.control
Plogcy5plogcy3slide110norm
dosR.WT_Replicate1
dosR.WT_Replicate4
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
Biological.replicate.1.of.7..panaxydol.treatment
Biological.replicate.3.of.6..negative.control
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.6.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.7.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DglnR.N_starvation_rep2
X2XR.rep2
Sub.MIC.INH
X4dbfcy3plogcy5slide903norm
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Biological.replicate.4.of.6..kanamycin.treatment
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
DglnR_without.vs..with.N_rep1
DglnR_without.vs..with.N_rep2
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.5.of.6..kanamycin.treatment
WT_stationary.phase_rep2
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
Biological.replicate.2.of.6..negative.control
Wild.type.rep1.x
Wild.type.rep1.y
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.7.of.7..ethambutol.treatment
Wild.type.rep3
Biological.replicate.4.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
mc2155.vs.Dcrp1.replicate.1
dcpA.knockout.rep2
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
X4XR.rep2
X4XR.rep1
Slow.vs..Fast.Replicate.4
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
Biological.replicate.5.of.6..negative.control
Biological.replicate.5.of.7..falcarinol.treatment
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.2.of.6..kanamycin.treatment
Comments
Log in to post comments