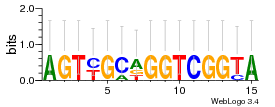
Module 616 Residual: 0.56
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.56 | 0.000073 | 39 | 82 | 22 | 20160204 |
| MSMEG_6911 ABC transporter, ATP-binding protein GluA | MSMEG_1854 Valosin containing protein-1 |
| MSMEG_6916 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.236) | MSMEG_1855 TPR repeat protein (Tetratricopeptide repeat protein, putative) |
| MSMEG_5670 Efflux membrane protein | MSMEG_6914 Uncharacterized protein |
| MSMEG_6691 Glutamine amidotransferase | MSMEG_3087 Preprotein translocase, SecG subunit (Protein-export membrane protein (Translocase subunit) SecG) |
| MSMEG_6690 Glutamine amidotransferase class-I (Peptidase C26) | MSMEG_3086 Triosephosphate isomerase (TIM) (EC 5.3.1.1) (Triose-phosphate isomerase) |
| MSMEG_3984 Transposase IS111A/IS1328/IS1533 (Transposase IS116/IS110/IS902 family protein) | MSMEG_3085 Phosphoglycerate kinase (EC 2.7.2.3) |
| MSMEG_6909 Amino acid ABC transporter, permease protein, 3-TM region, His/Glu/Gln/Arg/opine family protein (Polar amino acid ABC transporter, inner membrane subunit) (EC 4.2.1.51) | MSMEG_3084 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (EC 1.2.1.12) (NAD-dependent glyceraldehyde-3-phosphate dehydrogenase) |
| MSMEG_5668 Putative transcriptional regulator | MSMEG_6688 Purine catabolism PurC domain protein (Regulatory protein) |
| MSMEG_0957 Uncharacterized protein | MSMEG_2647 Metallophosphoesterase |
| MSMEG_0958 Uncharacterized protein | MSMEG_2648 4-phosphopantetheinyl transferase (Sfp-type phosphopantetheinyl transferase) |
| MSMEG_3344 Uncharacterized protein | MSMEG_2649 tRNA pseudouridine synthase B (EC 5.4.99.25) (tRNA pseudouridine(55) synthase) (tRNA pseudouridylate synthase) (tRNA-uridine isomerase) |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
KO_log.phase_rep1
Biological.replicate.5.of.7..panaxydol.treatment
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
dosR.WT_Replicate2
KO_log.phase_rep2
Wild.type.rep2.y
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.x
Wild.type.rep1.y
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep3
DglnR_without.vs..with.N_rep1
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DamtR.N.surplus_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_rep1
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.2.of.6..negative.control
X4XR.rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.with.vs..without.N_rep2
Biological.replicate.1.of.7..panaxydol.treatment
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.4.of.6..kanamycin.treatment
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
Comments
Log in to post comments