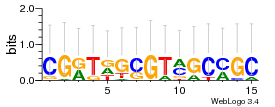
Module 629 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 1.5e-16 | 27 | 76 | 31 | 20160204 |
| MSMEG_1938 Uncharacterized protein | MSMEG_5216 Glyoxalase family protein (Glyoxalase/bleomycin resistance protein/dioxygenase) |
| MSMEG_5674 Major facilitator superfamily MFS_1 (Membrane transport protein) | MSMEG_6512 Acyl-CoA dehydrogenase (EC 1.3.8.-) (Acyl-CoA dehydrogenase domain protein) |
| MSMEG_5675 Pyridoxine/pyridoxamine 5'-phosphate oxidase (EC 1.4.3.5) (PNP/PMP oxidase) (PNPOx) (Pyridoxal 5'-phosphate synthase) | MSMEG_0289 Alpha/beta hydrolase fold family protein |
| MSMEG_4438 Uncharacterized protein | MSMEG_0440 Uncharacterized protein |
| MSMEG_0729 Uncharacterized protein | MSMEG_6229 Glycerol kinase (EC 2.7.1.30) |
| MSMEG_6016 Putative acyl-CoA dehydrogenase | MSMEG_6322 Diacylglycerol O-acyltransferase (EC 2.3.1.20) |
| MSMEG_6275 DNA polymerase III subunit epsilon (EC 2.7.7.7) | MSMEG_0248 Conserved transmembrane protein (Uncharacterized protein) |
| MSMEG_6014 Acyl-CoA dehydrogenase (EC 1.3.99.-) (Putative acyl-CoA dehydrogenase) | MSMEG_0249 Conserved transmembrane protein (Integral membrane protein) |
| MSMEG_5835 KsdD-like steroid dehydrogenase MSMEG_5835 (EC 1.3.99.-) | MSMEG_4213 Cytochrome P450 hydroxylase |
| MSMEG_6015 Acyl-CoA dehydrogenase FadE32 (Putative acyl-CoA dehydrogenase) | MSMEG_0245 Probable conserved transmembrane protein (Putative conserved transmembrane protein) |
| MSMEG_5837 Glutathione peroxidase | MSMEG_2811 Hydride transferase 1 (Luciferase family protein) |
| MSMEG_6231 Uncharacterized protein | MSMEG_0247 Secreted peptidase |
| MSMEG_2102 ABC transporter related protein (Nitrate transport ATP-binding protein NrtD) | MSMEG_4091 Nitrilotriacetate monooxygenase component A (EC 1.14.13.-) |
| MSMEG_0625 Uncharacterized protein | MSMEG_4093 Oxidoreductase, aldo/keto reductase family (Oxidoreductase, aldo/keto reductase family protein) |
| MSMEG_6622 Cytochrome P450 (Cytochrome P450 monooxygenase) | MSMEG_4092 Alkanal monooxygenase (Luciferase family protein) (EC 1.14.14.3) |
| MSMEG_6616 Alcohol dehydrogenase GroES-like protein (EC 1.1.1.284) (S-(Hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) |
Wild.type.rep1.y
KO_stationary.phase_rep1
Bedaquiline.vs..DMSO.30min
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
KO_log.phase_rep1
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate1
KO_log.phase_rep2
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.2.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.5
Biological.replicate.6.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
X3dbfcy3plogcy5slide607norm
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
dcpA.knockout.rep1
X4XR.rep1
X3dbfcy5plogcy3slide601norm
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
WT_log.phase_rep1
WT.with.vs..without.N_dye.swap_rep1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
mc2155.vs.Dcrp1.replicate.2
WT_stationary.phase_rep1
Biological.replicate.4.of.6..isoniazid.treatment
mc2155.vs.Dcrp1.replicate.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
WT.with.vs..without.N_dye.swap_rep2
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Biological.replicate.3.of.7..falcarinol.treatment
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.1.of.6..kanamycin.treatment
Comments
Log in to post comments