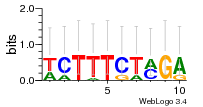
Module 630 Residual: 0.55
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.55 | 0.000000000000028 | 0.00014 | 76 | 24 | 20160204 |
| MSMEG_4601 Transcriptional regulator AraC family (Transcriptional regulator, AraC family protein) | MSMEG_4453 Uncharacterized protein |
| MSMEG_5553 CheR methyltransferase, SAM binding domain protein (EC 2.1.1.80) | MSMEG_4571 30S ribosomal protein S20 |
| MSMEG_3131 AMP-binding protein | MSMEG_3507 Fructose-bisphosphate aldolase (Fructose-bisphosphate aldolase class-I) (EC 4.1.2.13) |
| MSMEG_5710 Uncharacterized protein | MSMEG_4674 Trigger factor (TF) (EC 5.2.1.8) (PPIase) |
| MSMEG_4647 Carbohydrate kinase, PfkB, putative | MSMEG_1190 Anchored-membrane serine/threonine-protein kinase (EC 2.7.11.1) (Serine/threonine-protein kinase PknD) |
| MSMEG_1724 Uncharacterized protein | MSMEG_0323 Acyl-CoA dehydrogenase (EC 1.3.99.-) (Acyl-CoA dehydrogenase, short-chain specific, putative) |
| MSMEG_4013 Methylenetetrahydromethanopterin reductase (EC 1.5.99.11) (Oxidoreductase) | MSMEG_3552 Uncharacterized protein |
| MSMEG_6199 Transcriptional regulator WhiB | MSMEG_2527 Uncharacterized protein |
| MSMEG_6033 Uncharacterized protein | MSMEG_1118 Amino acid permease (Amino acid permease-associated region) |
| MSMEG_0339 FMN-dependent monooxygenase | MSMEG_1178 Transcriptional regulator |
| MSMEG_4186 Uncharacterized protein | MSMEG_1604 Cholesterol oxidase ChoD (EC 1.1.3.6) (FAD dependent oxidoreductase) |
| MSMEG_3061 Primosomal protein N | MSMEG_2811 Hydride transferase 1 (Luciferase family protein) |
KO_stationary.phase_rep2
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
Biological.replicate.4.of.6..negative.control
Bedaquiline.vs..DMSO.15min
Mycobacterium.smegmatis.Spheroplast.induced
Bedaquiline.vs..DMSO.30min
dosR.WT_Replicate3
dosR.WT_Replicate2
KO_log.phase_rep2
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
Wild.type.rep1.y
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.7..ethambutol.treatment
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.2
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
Biological.replicate.2.of.6..kanamycin.treatment
Biological.replicate.5.of.7..falcarinol.treatment
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy5plogcy3slide487norm
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.6.of.6..negative.control
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
Biological.replicate.3.of.6..kanamycin.treatment
WT.with.vs..without.N_dye.swap_rep1
X3dbfcy3plogcy5slide607norm
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
WT_stationary.phase_rep2
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Bedaquiline.vs..DMSO.60min
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments