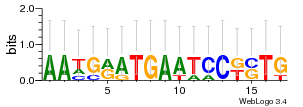
Module 639 Residual: 0.57
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.57 | 54 | 350 | 77 | 25 | 20160204 |
| MSMEG_5777 Transcription regulator | MSMEG_2204 Carveol dehydrogenase ((+)-trans-carveol dehydrogenase) (EC 1.1.1.275) (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.275) |
| MSMEG_4544 Methyltransferase small domain superfamily protein (Methyltransferase type 12) | MSMEG_4749 Uncharacterized protein |
| MSMEG_5323 Ornithine-acylacyl carrier protein N-acyltransferase (Uncharacterized protein) | MSMEG_4803 Cytochrome P450 (Cytochrome P450 superfamily protein) |
| MSMEG_0183 Uncharacterized protein | MSMEG_0778 Putative transcriptional regulator (Transcription regulator, GntR family) |
| MSMEG_6293 Transcriptional regulator, TetR family (Transcriptional regulatory protein) | MSMEG_0779 Short-chain dehydrogenase/reductase SDR |
| MSMEG_2894 Cyclohexanone monooxygenase (EC 1.14.13.22) (Steroid monooxygenase) | MSMEG_0630 Trehalose 2-sulfotransferase (EC 2.8.2.37) |
| MSMEG_4804 Uncharacterized protein | MSMEG_4660 Uncharacterized protein |
| MSMEG_2530 4-carboxymuconolactone decarboxylase domain protein (EC 4.1.1.44) | MSMEG_4886 Major facilitator family protein transporter (Major facilitator superfamily MFS_1) |
| MSMEG_6370 Alkyl hydroperoxide reductase AhpD (EC 1.11.1.15) | MSMEG_4332 Glycerol-3-phosphate dehydrogenase (EC 1.1.5.3) |
| MSMEG_3068 Uncharacterized protein | MSMEG_4333 Transcriptional regulator, TetR family (Transcriptional regulator, TetR family protein) |
| MSMEG_3573 Integral membrane protein (Integral membrane protein, putative) | MSMEG_4880 Methylmalonyl-CoA mutase C-terminal domain protein |
| MSMEG_2225 TetR family transcriptional regulator (Transcriptional regulator, TetR family protein) | MSMEG_6587 ATP-dependent helicase HrpA |
| MSMEG_4064 Zinc-binding alcohol dehydrogenase |
Bedaquiline.vs..DMSO.30min
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
Biological.replicate.4.of.6..negative.control
Biological.replicate.3.of.7..ethambutol.treatment
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate2
Biological.replicate.4.of.6..kanamycin.treatment
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Plogcy5plogcy3slide110norm
X3dbfcy5plogcy3slide601norm
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
MB100pruR.vs..MB100.rep.2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Bedaquiline.vs..DMSO.60min
rel.knockout.rep2
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.7..falcarinol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DamtR.N.surplus_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
stphcy5plogcy3slide487norm
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
stphcy3plogcy5slide581norm
WT_log.phase_rep1
DglnR_without.vs..with.N_rep1
X4dbfcy5plogcy3slide480norm
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
X2.5..vs..50..air.saturation.Replicate.3
Wild.type.rep1.x
Biological.replicate.1.of.7..ethambutol.treatment
X4XR.rep2
Wild.type.rep2.x
Slow.vs..Fast.Replicate.5
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X3dbfcy5plogcy3slide603norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
X2XR.rep1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.vs..DglnR.N_starvation_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments