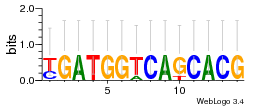
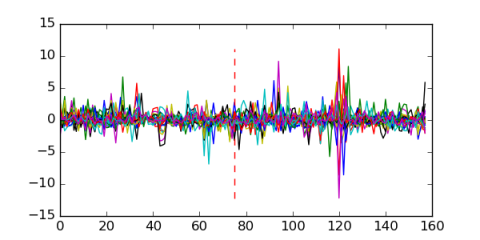
Module 647 Residual: 0.57
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.57 | 0.000000062 | 0.00046 | 75 | 26 | 20160204 |
| MSMEG_5328 Uncharacterized protein | MSMEG_6835 Fatty acid desaturase (EC 1.14.19.3) |
| MSMEG_4135 Possible lysine decarboxylase superfamily protein (Uncharacterized protein) | MSMEG_3234 Integral membrane protein |
| MSMEG_0291 Dioxygenase, TauD/TfdA family protein (Taurine catabolism dioxygenase TauD/TfdA) | MSMEG_3235 ABC-type amino acid transport system, secreted component (Extracellular solute-binding protein family 3) |
| MSMEG_3553 D-hydantoinase (Dihydropyrimidinase) (Dihydropyrimidinase) (EC 3.5.2.2) | MSMEG_3236 ABC-type amino acid transport system, permease component (Truncated polar amino acid ABC transporter) |
| MSMEG_3554 Methylenetetrahydromethanopterin reductase (EC 1.5.99.11) (N5,N10-methylene-tetrahydromethanopterin reductase) | MSMEG_3237 ATP-binding protein |
| MSMEG_3713 Uncharacterized protein | MSMEG_2526 Amine oxidase (EC 1.4.3.-) |
| MSMEG_3711 Uncharacterized protein | MSMEG_1775 Cytochrome P450 (Cytochrome P450 monooxygenase) |
| MSMEG_6833 Alcohol dehydrogenase (EC 1.1.1.1) | MSMEG_6722 Uncharacterized protein |
| MSMEG_6832 Uncharacterized protein | MSMEG_5069 Sec-independent protein translocase protein TatB |
| MSMEG_5070 Serine protease htrA (Trypsin) | MSMEG_5068 ATP-binding Mrp protein (Mrp protein) |
| MSMEG_5071 Anti-sigma-E factor RseA (Regulator of SigE) (Sigma-E anti-sigma factor RseA) | MSMEG_4099 ABC transporter permease protein (Binding-protein-dependent transport systems inner membrane component) |
| MSMEG_4101 ABC-type transporter, substrate binding protein | MSMEG_4098 ABC transporter ATP-binding protein |
| MSMEG_4100 ABC transporter permease protein (ABC-type transport systems, permease component) | MSMEG_5067 Putative membrane protein (Secreted protein) |
KO_stationary.phase_rep2
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
Biological.replicate.3.of.6..kanamycin.treatment
Bedaquiline.vs..DMSO.15min
dosR.WT_Replicate4
Biological.replicate.4.of.6..isoniazid.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate1
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep2
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.x
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
Wild.type.rep3
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
X3dbfcy5plogcy3slide601norm
DglnR_without.vs..with.N_rep1
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
X4dbfcy5plogcy3slide901norm
dcpA.knockout.rep2
X4dbfcy5plogcy3slide480norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.2.of.6..isoniazid.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X3dbfcy5plogcy3slide603norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
Wild.type.rep2.x
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate2
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments