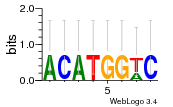
Module 677 Residual: 0.41
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.41 | 11 | 470 | 85 | 19 | 20160204 |
| MSMEG_2063 NADH-quinone oxidoreductase subunit A (EC 1.6.5.11) (NADH dehydrogenase I subunit A) (NDH-1 subunit A) (NUO1) | MSMEG_2059 NADH-quinone oxidoreductase chain e (EC 1.6.99.5) |
| MSMEG_2060 NADH-quinone oxidoreductase subunit D (EC 1.6.5.11) (NADH dehydrogenase I subunit D) (NDH-1 subunit D) | MSMEG_2058 NADH-quinone oxidoreductase subunit F (EC 1.6.99.5) (NADH-quinone oxidoreductase, F subunit) (EC 1.6.99.5) |
| MSMEG_2061 NADH-quinone oxidoreductase subunit C (EC 1.6.5.11) (NADH dehydrogenase I subunit C) (NDH-1 subunit C) | MSMEG_0244 Two component response transcriptional regulatory protein prra |
| MSMEG_2230 Amidohydrolase 2 (Amidohydrolase family protein) | MSMEG_2056 NADH-quinone oxidoreductase subunit H (EC 1.6.5.11) (NADH dehydrogenase I subunit H) (NDH-1 subunit H) |
| MSMEG_2051 NADH-quinone oxidoreductase, M subunit (EC 1.6.99.5) | MSMEG_2055 NADH-quinone oxidoreductase subunit I (EC 1.6.5.11) (NADH dehydrogenase I subunit I) (NDH-1 subunit I) |
| MSMEG_2052 NADH-quinone oxidoreductase chain l (NADH-quinone oxidoreductase, L subunit) (EC 1.6.99.5) | MSMEG_2053 NADH-quinone oxidoreductase subunit K (EC 1.6.5.11) (NADH dehydrogenase I subunit K) (NDH-1 subunit K) |
| MSMEG_2231 AMP-binding enzyme (Putative acyl-CoA synthetase, long-chain fatty acid:CoA ligase) (EC 2.3.1.86) | MSMEG_0246 Sensor-type histidine kinase PrrB (Sensor-type histidine kinase prrB) (EC 2.7.13.3) |
| MSMEG_2234 Acyl-CoA dehydrogenase (EC 1.3.8.-) (Acyl-CoA dehydrogenase, middle domain protein) | MSMEG_2229 Enoyl-CoA hydratase (Enoyl-CoA hydratase/isomerase) (EC 4.2.1.17) |
| MSMEG_2233 Acyl-CoA dehydrogenase (EC 1.3.8.-) (Acyl-CoA dehydrogenase, C-domain protein) | MSMEG_0243 Uncharacterized protein |
| MSMEG_2232 Amidohydrolase 2 (Amidohydrolase family protein) |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DglnR.N_starvation_rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
X4XR.rep1
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.4.of.6..negative.control
Plogcy5plogcy3slide110norm
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.5.of.6..negative.control
Biological.replicate.2.of.6..kanamycin.treatment
KO_log.phase_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.x
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
rel.knockout.rep2
Wild.type.rep3
Biological.replicate.1.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep1
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
WT.vs..DamtR.N.surplus_rep1
stphcy5plogcy3slide487norm
Wild.type.rep2.y
mc2155.vs.Dcrp1.replicate.1
X3dbfcy3plogcy5slide607norm
Biological.replicate.3.of.6..negative.control
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.2.of.7..falcarinol.treatment
stphcy5plogcy3slide580norm
WT_log.phase_rep2
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
X4dbfcy5plogcy3slide480norm
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.1
WT_stationary.phase_rep2
WT_stationary.phase_rep1
X4XR.rep2
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
X4dbfcy5plogcy3slide901norm
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
Wild.type.rep2.x
X2XR.rep2
Biological.replicate.1.of.7..panaxydol.treatment
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.2.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments