MSMEG_0935 MSMEG_0935 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (BPG-dependent PGAM) (PGAM) (Phosphoglyceromutase) (dPGM) (EC 5.4.2.11)
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Locus Tag | Symbol | Synonyms | Product | Protein Synonyms | Start | End | Strand | PubMed ID | |
|---|---|---|---|---|---|---|---|---|---|
| MSMEG_0935 | MSMEG_0935 | gpmA | gpmA, MSMEG_0935, MSMEI_0912 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (BPG-dependent PGAM) (PGAM) (Phosphoglyceromutase) (dPGM) (EC 5.4.2.11) | 17295914, 18955433 |
| Residual | Motif 1 | Expression Plot | |
|---|---|---|---|
| msm_bicluster_0113 | 0.57 |
 |
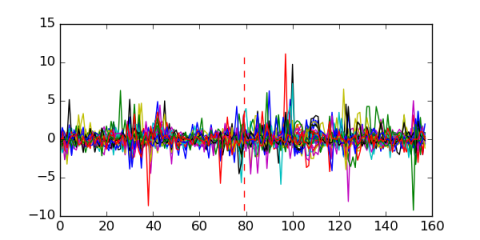 |
| msm_bicluster_0323 | 0.58 |
 |
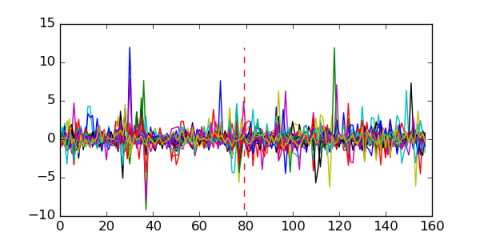 |
| Uniprot | NCBI Gene | NCBI Protein | EnsemblBacteria | InterPro | EggNOG | OrthoDB |
|---|---|---|---|---|---|---|
| A0QR00 | 4537124; | WP_003892361.1, YP_885338.1 | ABK73234, AFP37392, AIU06191 | IPR013078, IPR029033, IPR001345, IPR005952 | COG0588, ENOG4107RPC | EOG6C8N1H |
| Title | Insert | TAG Module |
|---|---|---|
| NA |
| Orthologues | Paralogues |
|---|---|
| Catalytic Activity | KEGG Ontology | Pathways |
|---|---|---|
| 2-phospho-D-glycerate = 3-phospho-D-glycerate, ECO:0000256|HAMAP-Rule:MF_01039, ECO:0000256|RuleBase:RU004512, ECO:0000256|SAAS:SAAS00366459}. | msb:LJ00_04640, msg:MSMEI_0912, msm:MSMEG_0935 | Carbohydrate degradation, glycolysis, pyruvate from D-glyceraldehyde 3-phosphate: step 3/5. {ECO:0000256|HAMAP-Rule:MF_01039, ECO:0000256|RuleBase:RU004512}. |
| Product (LegacyBRC) | Product (RefSeq) |
|---|---|
| GI Number | Accession | Blast | Conserved Domains |
|---|---|---|---|
| Link to STRINGS | STRINGS Network |
|---|---|
| MSMEG_0935 |
Add comment
Log in to post comments