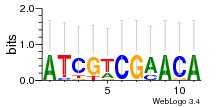
Module 127 Residual: 0.58
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.58 | 3.4e-32 | 0.000000000088 | 79 | 30 | 20160204 |
| MSMEG_6487 2-hydroxy-3-carboxy-6-oxo-7-methylocta-2, 4-dienoate decarboxylase (Aminocarboxymuconate-semialdehyde decarboxylase) (EC 4.1.1.45) | MSMEG_0114 Extracellular solute-binding protein, family protein 3 (Substrate-binding region of ABC-type glycine betaine transport system) |
| MSMEG_1492 MarR-transcriptional regulator (Transcriptional regulator, MarR family protein) | MSMEG_1325 RecBCD enzyme subunit RecD (EC 3.1.11.5) (Exonuclease V subunit RecD) (ExoV subunit RecD) (Helicase/nuclease RecBCD subunit RecD) |
| MSMEG_3261 Uncharacterized protein | MSMEG_0116 ABC transporter related protein (Taurine import ATP-binding protein TauB) (EC 3.6.3.36) |
| MSMEG_4756 Holo-[acyl-carrier-protein] synthase (Holo-ACP synthase) (EC 2.7.8.7) (4'-phosphopantetheinyl transferase AcpS) | MSMEG_1327 RecBCD enzyme subunit RecB (EC 3.1.11.5) (Exonuclease V subunit RecB) (ExoV subunit RecB) (Helicase/nuclease RecBCD subunit RecB) |
| MSMEG_4757 Fatty acid synthase | MSMEG_0111 Putative magnesium transport protein |
| MSMEG_3441 Uncharacterized protein | MSMEG_0112 Uncharacterized protein |
| MSMEG_6465 Chloride channel (Voltage-gated chloride channel) | MSMEG_0113 Taurine transport system permease protein TauC |
| MSMEG_5349 Uncharacterized protein | MSMEG_3650 Integral membrane protein |
| MSMEG_4759 Uncharacterized protein | MSMEG_6872 Beta-lactamase (Uncharacterized protein) |
| MSMEG_2792 ATP-dependent protease ATP-binding subunit ClpC2 (Clp amino terminal domain protein) | MSMEG_4520 Sulfate/thiosulfate ABC transporter, ATP-binding protein (EC 3.6.3.25) (Sulfate/thiosulfate import ATP-binding protein CysA) |
| MSMEG_2793 Sensor-type histidine kinase PrrB (EC 2.7.13.3) | MSMEG_4962 Monooxygenase FAD-binding protein (RemO protein) |
| MSMEG_3966 Uncharacterized protein | MSMEG_3442 Cyclohexadienyl dehydratase (EC 4.2.1.-) (EC 4.2.1.51) (EC 4.2.1.91) |
| MSMEG_0174 Putative inner membrane protein (Uncharacterized protein) | MSMEG_4988 Integral membrane protein |
| MSMEG_0175 FAD dependent oxidoreductase, putative (Fructosyl-amino acid oxidase, putative) | MSMEG_1182 Imidazolonepropionase (EC 3.5.2.7) (Imidazolone-5-propionate hydrolase) |
| MSMEG_1328 RecBCD enzyme subunit RecC (EC 3.1.11.5) (Exonuclease V subunit RecC) (ExoV subunit RecC) (Helicase/nuclease RecBCD subunit RecC) | MSMEG_1183 Histidine ammonia-lyase (EC 4.3.1.3) |
Wild.type.rep1.y
mc2155.vs.Dcrp1.replicate.1
WT.vs..DamtR.without.N_rep1
KO_log.phase_rep1
X4XR.rep1
Bedaquiline.vs..DMSO.15min
Wild.type.rep2.x
Wild.type.rep3
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.2.of.6..negative.control
KO_log.phase_rep2
Wild.type.rep2.y
DglnR_without.vs..with.N_rep1
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.3.of.6..kanamycin.treatment
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
M..smegmatis.Acid.NO.treated
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.6..kanamycin.treatment
Sub.MIC.INH
X0.6..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..falcarinol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.7.of.7..panaxydol.treatment
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
dcpA.knockout.rep1
Plogcy5plogcy3slide111norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
WT_stationary.phase_rep1
X4XR.rep2
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.4
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Slow.vs..Fast.Replicate.3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DglnR.N_starvation_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.1.of.6..kanamycin.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Comments
Log in to post comments