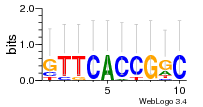
Module 141 Residual: 0.42
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.42 | 0.0000000013 | 0.00000014 | 84 | 26 | 20160204 |
| MSMEG_0849 Oxygenase | MSMEG_4169 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100) (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_0845 Multisubunit Na+/H+ antiporter (Multisubunit Na+/H+ antiporter MnhE subunit-like protein) | MSMEG_4821 Uncharacterized protein |
| MSMEG_6733 Hydrolase, carbon-nitrogen family protein | MSMEG_5317 Conserved membrane protein (Uncharacterized protein) |
| MSMEG_6507 Glycogen operon protein GlgX homolog (EC 3.2.1.-) | MSMEG_5566 Transcriptional regulator |
| MSMEG_6391 Propionyl-CoA carboxylase beta chain (EC 6.4.1.3) (Propionyl-CoA carboxylase beta chain 4 accD4) | MSMEG_1835 TobH protein |
| MSMEG_6392 Polyketide synthase | MSMEG_4167 Alcohol dehydrogenase B (EC 1.1.1.1) (Zinc-type alcohol dehydrogenase AdhD) (EC 1.1.1.1) |
| MSMEG_6393 Acyl-CoA synthase (Fatty-acid-CoA ligase FadD32) | MSMEG_4166 Acyl-CoA dehydrogenase (EC 1.3.8.4) |
| MSMEG_0829 Exodeoxyribonuclease III (EC 3.1.11.2) (Exodeoxyribonuclease III protein XthA) (EC 3.1.11.2) | MSMEG_5315 Gp35 protein |
| MSMEG_4951 50S ribosomal protein L31 | MSMEG_0148 Transcriptional regulator, TetR family protein |
| MSMEG_1452 Sulfatase-modifying factor 1 | MSMEG_0678 Deoxycytidine triphosphate deaminase (dCTP deaminase) (EC 3.5.4.13) |
| MSMEG_1451 Arylsulfatase | MSMEG_4168 Acetate CoA-transferase YdiF (EC 2.8.3.8) |
| MSMEG_1784 Topoisomerase IB (EC 5.99.1.2) (Type I topoisomerase) | MSMEG_2914 L-idonate 5-dehydrogenase (EC 1.1.1.264) |
| MSMEG_0843 Monovalent cation/proton antiporter, MnhG/PhaG subunit (Multisubunit Na+/H+ antiporter, G subunit) | MSMEG_2912 Inner membrane metabolite transport protein YdfJ |
Wild.type.rep1.y
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
KO_log.phase_rep1
Biological.replicate.4.of.6..negative.control
WT.vs..DamtR.without.N_rep2
Biological.replicate.3.of.7..ethambutol.treatment
Mycobacterium.smegmatis.Spheroplast.induced
X3dbfcy5plogcy3slide603norm
Biological.replicate.2.of.7..falcarinol.treatment
Biological.replicate.5.of.6..kanamycin.treatment
dosR.WT_Replicate1
KO_log.phase_rep2
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.60min
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.2.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
mc2155.vs.Dcrp1.replicate.2
Plogcy5plogcy3slide113norm
Slow.vs..Fast.Replicate.3
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
DglnR_without.vs..with.N_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..negative.control
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.4.of.7..falcarinol.treatment
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.N.surplus_dyeswap_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Comments
Log in to post comments