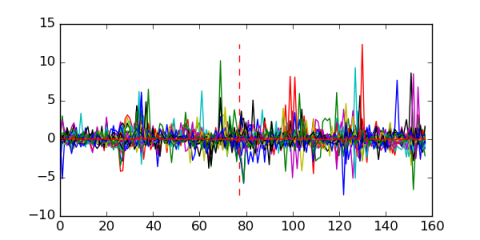
Module 156 Residual: 0.56
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.56 | 1.9e-32 | 0.0000000047 | 77 | 31 | 20160204 |
| MSMEG_3351 Uncharacterized protein | MSMEG_1329 Major facilitator superfamily MFS_1 (Major facilitator superfamily protein MFS_1, putative) |
| MSMEG_1864 Integrase catalytic region (Transposase) | MSMEG_3063 LemA protein |
| MSMEG_5409 Uncharacterized protein | MSMEG_5096 |
| MSMEG_0432 Uroporphyrinogen III synthase HEM4 (EC 4.2.1.75) (Uroporphyrinogen-III synthetase) | MSMEG_5097 Uncharacterized protein |
| MSMEG_0431 Cobalamin (Vitamin B12) biosynthesis CbiX protein (EC 4.99.1.3) (Secreted protein) | MSMEG_1730 IS629 transposase orfB (ISMsm1, transposase orfB) |
| MSMEG_6778 Uncharacterized protein | MSMEG_5825 Na+/H+ antiporter (NhaP-type Na+/H+ and K+/H+ antiporters with a unique C-terminal domain protein) |
| MSMEG_4870 FAD binding domain, putative (Fumarate reductase/succinate dehydrogenase flavoprotein-like protein) | MSMEG_1584 Uncharacterized protein |
| MSMEG_0439 UPF0324 membrane protein (Uncharacterized protein) | MSMEG_0440 Uncharacterized protein |
| MSMEG_0438 Periplasmic binding protein (Periplasmic iron-transport lipoprotein) | MSMEG_2819 |
| MSMEG_1330 MarR-family protein transcriptional regulator | MSMEG_2818 |
| MSMEG_2631 DNA-damage-inducible protein F DinF (MATE efflux family protein) | MSMEG_2628 Translation initiation factor IF-2 |
| MSMEG_2630 DHH family protein | MSMEG_2629 Ribosome-binding factor A |
| MSMEG_0490 Enoyl-CoA hydratase (Enoyl-CoA hydratase/isomerase family protein) (EC 4.2.1.17) | MSMEG_1213 Cytochrome P450 monooxygenase |
| MSMEG_1162 Nitrilotriacetate monooxygenase component A (EC 1.14.13.-) (Nitrilotriacetate monooxygenase component A (NTA monooxygenase component A) (NTA-MO A)) | MSMEG_3915 NAD-dependent alcohol dehydrogenase (EC 1.1.1.1) |
| MSMEG_6851 PEBP family protein (Phosphatidylethanolamine-binding protein) | MSMEG_0489 Putative recemase (Racemase) |
| MSMEG_5115 Uncharacterized protein |
KO_stationary.phase_rep2
Biological.replicate.3.of.6..kanamycin.treatment
M..smegmatis.Acid.NO.treated
dosR.WT_Replicate4
Biological.replicate.1.of.6..negative.control
Biological.replicate.5.of.6..kanamycin.treatment
dosR.WT_Replicate2
dosR.WT_Replicate1
KO_log.phase_rep2
Biological.replicate.2.of.7..panaxydol.treatment
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
WT.vs..DamtR.N.surplus_rep1
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep1
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
DglnR_without.vs..with.N_rep1
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
X4XR.rep1
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Bedaquiline.vs..DMSO.60min
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.3
X3dbfcy5plogcy3slide603norm
mc2155.vs.Dcrp1.replicate.1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.5.of.7..falcarinol.treatment
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
Wild.type.rep1.y
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Sub.MIC.INH
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.4.of.6..kanamycin.treatment
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments