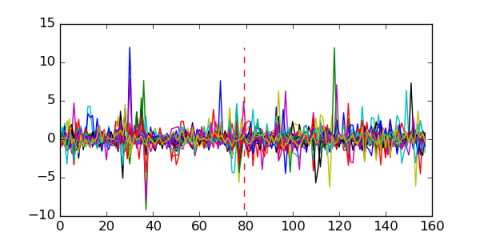
Module 323 Residual: 0.58
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.58 | 0.0000000000037 | 0.00000088 | 79 | 27 | 20160204 |
| MSMEG_4600 Uncharacterized protein | MSMEG_6072 Arsenical pump membrane protein |
| MSMEG_6096 Type III pantothenate kinase (EC 2.7.1.33) (PanK-III) (Pantothenic acid kinase) | MSMEG_0884 Glyoxalase family protein (Glyoxalase/bleomycin resistance protein/dioxygenase) |
| MSMEG_4602 Uncharacterized protein | MSMEG_3305 Integral membrane protein DUF6 (Uncharacterized protein) |
| MSMEG_3527 Putative HTH-type transcriptional regulator | MSMEG_4024 Transcriptional regulator, TetR family protein |
| MSMEG_3222 Prolipoprotein diacylglyceryl transferase (EC 2.4.99.-) | MSMEG_5373 Nitrilase 2 (EC 3.5.5.1) |
| MSMEG_3221 Tryptophan synthase alpha chain (EC 4.2.1.20) | MSMEG_3218 Trp region conserved hypothetical membrane protein |
| MSMEG_3220 Tryptophan synthase beta chain (EC 4.2.1.20) | MSMEG_3219 Indole-3-glycerol phosphate synthase (IGPS) (EC 4.1.1.48) |
| MSMEG_3201 Nicotinate-nucleotide pyrophosphorylase (EC 2.4.2.19) | MSMEG_0935 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (BPG-dependent PGAM) (PGAM) (Phosphoglyceromutase) (dPGM) (EC 5.4.2.11) |
| MSMEG_3200 L-aspartate oxidase (EC 1.4.3.16) | MSMEG_3216 Peroxidoxin BcpB (EC 1.11.1.15) (Peroxiredoxin Q) |
| MSMEG_5594 Ferredoxin-dependent glutamate synthase (EC 1.4.1.13) | MSMEG_3217 Anthranilate synthase component 1 (AS) (ASI) (EC 4.1.3.27) |
| MSMEG_5593 Pyruvate dehydrogenase (Thiamine pyrophosphate enzyme-like TPP binding protein) (EC 1.2.3.3) | MSMEG_1755 Anti-sigm factor, ChrR |
| MSMEG_3199 Quinolinate synthase (EC 2.5.1.72) | MSMEG_0931 Uncharacterized protein |
| MSMEG_3198 Hydrolase, NUDIX family protein | MSMEG_1234 ABC transporter ATP-binding protein (Taurine import ATP-binding protein TauB) (EC 3.6.3.36) |
| MSMEG_0883 Amidohydrolase family protein (Amidohydrolase family protein/D-aminoacylase domain protein) (EC 3.5.2.3) |
KO_stationary.phase_rep1
Biological.replicate.5.of.7..falcarinol.treatment
Wild.type.rep2.y
WT.vs..DamtR.without.N_rep2
Biological.replicate.4.of.6..negative.control
X4XR.rep1
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.3.of.7..ethambutol.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate2
stphcy5plogcy3slide487norm
KO_log.phase_rep1
Wild.type.rep1.x
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
mc2155.vs.Dcrp1.replicate.3
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
mc2155.vs.Dcrp1.replicate.1
X3dbfcy3plogcy5slide607norm
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
Biological.replicate.2.of.6..isoniazid.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
WT_log.phase_rep1
WT.with.vs..without.N_dye.swap_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.7.of.7..ethambutol.treatment
WT_stationary.phase_rep1
X4XR.rep2
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Biological.replicate.5.of.7..ethambutol.treatment
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Wild.type.rep3
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
Biological.replicate.1.of.6..negative.control
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.2.of.7..ethambutol.treatment
WT.vs..DamtR.N.surplus_rep2
WT.vs..DglnR.N_starvation_dye.swap_rep1
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments