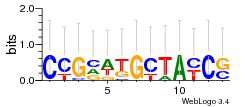
Module 391 Residual: 0.47
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.47 | 5.2e-19 | 0.016 | 83 | 22 | 20160204 |
| MSMEG_3223 TM2 domain containing protein+B7201 (TM2 domain protein) | MSMEG_4263 Ubiquinol-cytochrome C reductase QcrB (EC 1.10.2.2) (Ubiquinol-cytochrome c reductase cytochrome b subunit) |
| MSMEG_3224 Conserved membrane protein (Uncharacterized protein) | MSMEG_4260 Cytochrome c oxidase subunit 3 (EC 1.9.3.1) |
| MSMEG_2318 | MSMEG_4261 Ubiquinol-cytochrome c reductase cytochrome c subunit |
| MSMEG_2319 | MSMEG_3148 Uncharacterized protein |
| MSMEG_3229 Uncharacterized protein | MSMEG_3149 UPF0353 protein MSMEG_3149 |
| MSMEG_1666 Alternative RNA polymerase sigma factor SigJ (RNA polymerase sigma-70 factor) | MSMEG_3147 ATPase, MoxR family protein |
| MSMEG_1668 Pyridoxamine 5'-phosphate oxidase family protein | MSMEG_3231 ABC transporter, CydDC cysteine exporter (CydDC-E) family protein, permease/ATP-binding protein CydD (Transmembrane ATP-binding protein ABC transporter cydD) (EC 3.6.3.25) |
| MSMEG_4268 Cytochrome c oxidase subunit 2 (EC 1.9.3.1) (Transmembrane cytochrome C oxidase (Subunit II) ctaC (Transmembrane cytochrome C oxidase subunit II ctaC)) | MSMEG_3232 Cytochrome D ubiquinol oxidase, subunit II (EC 1.10.3.-) (Integral membrane cytochrome D ubiquinol oxidase (Subunit II) CydB) (EC 3.1.3.1) |
| MSMEG_4269 Asparagine synthase (Glutamine-hydrolyzing) (EC 6.3.5.4) (Asparagine synthetase AsnB) | MSMEG_3233 Cytochrome D ubiquinol oxidase subunit 1 (EC 1.10.3.-) (Putative cytochrome ubiquinol oxidase subunit) |
| MSMEG_4267 Cytochrome c oxidase polypeptide 4 (EC 1.9.3.1) (Cytochrome aa3 subunit 4) (Cytochrome c oxidase polypeptide IV) | MSMEG_1039 FEIII-dicitrate-binding periplasmic lipoprotein FecB (Periplasmic binding protein) |
| MSMEG_4262 Ubiquinol-cytochrome c reductase iron-sulfur subunit | MSMEG_4258 Anthranilate phosphoribosyltransferase (EC 2.4.2.18) |
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
Biological.replicate.4.of.6..negative.control
Sub.MIC.INH
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.5.of.6..kanamycin.treatment
KO_log.phase_rep2
DglnR_without.vs..with.N_rep1
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.6.of.7..ethambutol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Bedaquiline.vs..DMSO.60min
Biological.replicate.3.of.7..falcarinol.treatment
X4XR.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.5.of.7..panaxydol.treatment
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
Biological.replicate.3.of.6..kanamycin.treatment
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
Biological.replicate.1.of.7..ethambutol.treatment
X4XR.rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
X3dbfcy5plogcy3slide603norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Comments
Log in to post comments