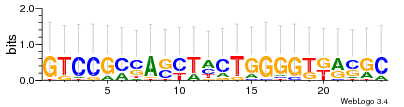
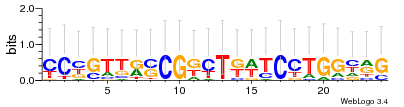
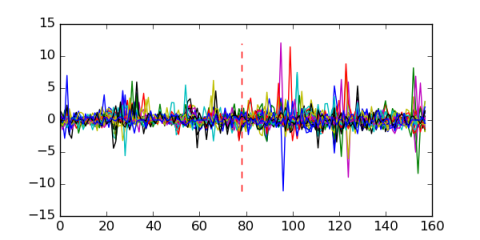
Module 468 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 0.00000000000011 | 0.00000026 | 78 | 29 | 20160204 |
| MSMEG_4359 Uncharacterized protein | MSMEG_0281 Choline dehydrogenase (Glucose-methanol-choline oxidoreductase) (EC 1.1.99.1) |
| MSMEG_3687 AMP-dependent synthetase and ligase (EC 2.3.1.86) (Medium chain acyl-CoA synthetase) | MSMEG_0282 Putative ethyl tert-butyl ether degradation protein EthD (Uncharacterized protein) |
| MSMEG_3011 Cyclohexanone 1,2-monooxygenase (EC 1.14.13.22) (Steroid monooxygenase) | MSMEG_5455 Pe family protein |
| MSMEG_0429 Putative ferric uptake regulator | MSMEG_4696 Alpha-amylase family protein (Putative alpha-glucosidase) (EC 3.2.1.20) |
| MSMEG_6118 IclR-family protein transcriptional regulator (IclR-family transcriptional regulator) | MSMEG_0280 Alpha/beta hydrolase fold (Alpha/beta hydrolase fold-3) |
| MSMEG_4952 Acyl-CoA synthase (Fatty-acid-CoA ligase FadD1) | MSMEG_4722 Short-chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_4953 Putative transcriptional regulator | MSMEG_2170 3-hydroxyisobutyrate dehydrogenase family protein |
| MSMEG_2390 Hydrolase, NUDIX family protein | MSMEG_6110 Hypoxanthine phosphoribosyltransferase (EC 2.4.2.8) |
| MSMEG_2391 Polyphosphate kinase (EC 2.7.4.1) | MSMEG_2760 Polyphosphate glucokinase (Polyphosphate glucokinase PpgK) (EC 2.7.1.63) |
| MSMEG_2168 Uncharacterized protein | MSMEG_2762 Inositol-1-monophosphatase (EC 3.1.3.25) |
| MSMEG_3909 Mycobacterium tuberculosis paralogous family 11 (Uncharacterized protein) | MSMEG_2872 Short-chain dehydrogenase/reductase SDR |
| MSMEG_4309 Low molecular weight protein-tyrosine-phosphatase (EC 3.1.3.48) (Phosphotyrosine protein phosphatase PtpA) | MSMEG_3666 C-5 sterol desaturase (EC 1.3.-.-) |
| MSMEG_4308 5'-nucleotidase (EC 3.1.3.5) (HAD-superfamily hydrolase, subfamily IA, variant 1) (EC 3.1.3.18) | MSMEG_2873 3-ketosteroid-delta4 (Uncharacterized protein) |
| MSMEG_4721 Permease of the major facilitator superfamily protein | MSMEG_4311 Conserved transmembrane protein (Uncharacterized protein) |
| MSMEG_5456 LpqN (MK35 lipoprotein) |
Plogcy5plogcy3slide113norm
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
dcpA.knockout.rep2
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.2.of.7..falcarinol.treatment
dosR.WT_Replicate3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
Wild.type.rep1.y
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
X4XR.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Wild.type.rep2.y
mc2155.vs.Dcrp1.replicate.1
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.5.of.7..panaxydol.treatment
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.2.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
WT_log.phase_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
WT.with.vs..without.N_dye.swap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X3dbfcy5plogcy3slide603norm
X4XR.rep2
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
WT.vs..DglnR.N_starvation_dye.swap_rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
X4dbfcy5plogcy3slide480norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.vs..DglnR.N_starvation_rep2
X2XR.rep1
X2XR.rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments