Module 365 Residual: 0.53
| Title | Bicluster Model | Residual | Score |
|---|---|---|---|
| bicluster_0365 | v02 | 0.53 | -9.64 |
| Rv0057 | Rv1475c Aconitate hydratase (EC 4.2.1.3) @ 2-methylisocitrate dehydratase (EC 4.2.1.99) |
| Rv1637c metallo-beta-lactamase superfamily protein | Rv2431c PE family protein |
| Rv3367 PE_PGRS family protein | Rv0011c FIG018426: putative septation inhibitor protein |
| Rv0020c | Rv0192 YkuD domain-containing hypothetical lipoprotein |
| Rv0469 POSSIBLE MYCOLIC ACID SYNTHASE UMAA1 | Rv0559c POSSIBLE CONSERVED SECRETED PROTEIN |
| Rv0909 | Rv0951 Succinyl-CoA ligase [ADP-forming] beta chain (EC 6.2.1.5) |
| Rv1174c LOW MOLECULAR WEIGHT T-CELL ANTIGEN TB8.4 | Rv1749c Possible membrane protein |
| Rv1750c Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3) | Rv2773c Dihydrodipicolinate reductase (EC 1.3.1.26) |
|
GO:0003994 | aconitate hydratase activity |
|
GO:0005576 | extracellular region |
|
GO:0005618 | cell wall |
|
GO:0005829 | cytosol |
|
GO:0005886 | plasma membrane |
|
GO:0010039 | response to iron ion |
|
GO:0030350 | iron-responsive element binding |
|
GO:0040007 | growth |
|
GO:0005515 | protein binding |
|
GO:0071768 | mycolic acid biosynthetic process |
|
GO:0045927 | positive regulation of growth |
|
GO:0004775 | succinate-CoA ligase (ADP-forming) activity |
|
GO:0008839 | dihydrodipicolinate reductase activity |
|
GO:0019877 | diaminopimelate biosynthetic process |
|
GO:0051260 | protein homooligomerization |
|
GO:0070404 | NADH binding |



















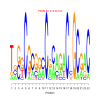
Discussion