Genes
Gene annotations were collated from multiple sources;
- MaGe MicroScope Publication: PMID:21349987
- Uniprot
- PATRIC
Gene essentiality is based on TnSeq Data from: Dembek M, Barquist L, Boinett CJ, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6(2):e02383. Published 2015 Feb 24.doi:10.1128/mBio.02383-14
| Title | Short Name | Product | Essentiality(TnSeq) | Essentiality(in vivo) | Essentiality(Rich Broth) | Expression | |
|---|---|---|---|---|---|---|---|
| CD630_00010 | dnaA | Chromosomal replication initiator protein |
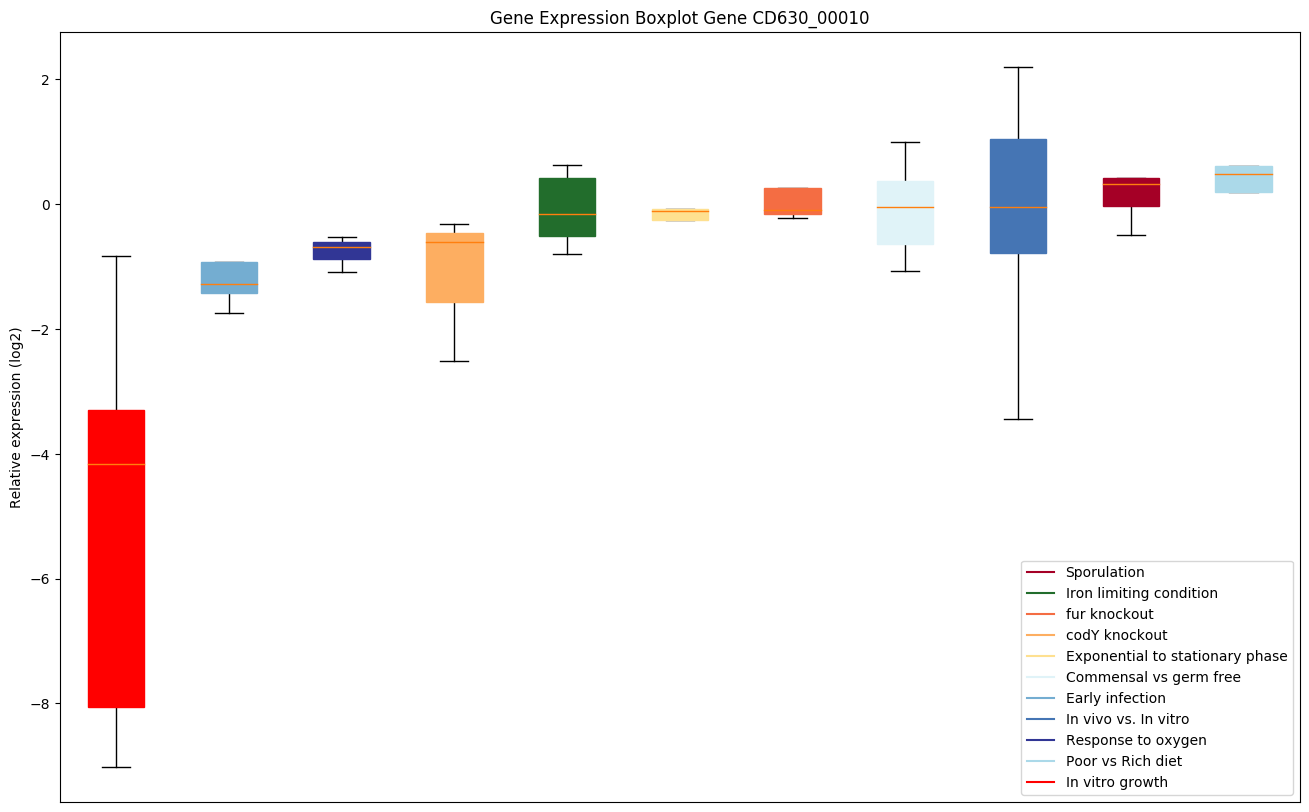 |
Bookmark | |||
| CD630_00020 | dnaN | DNA polymerase III subunit beta |
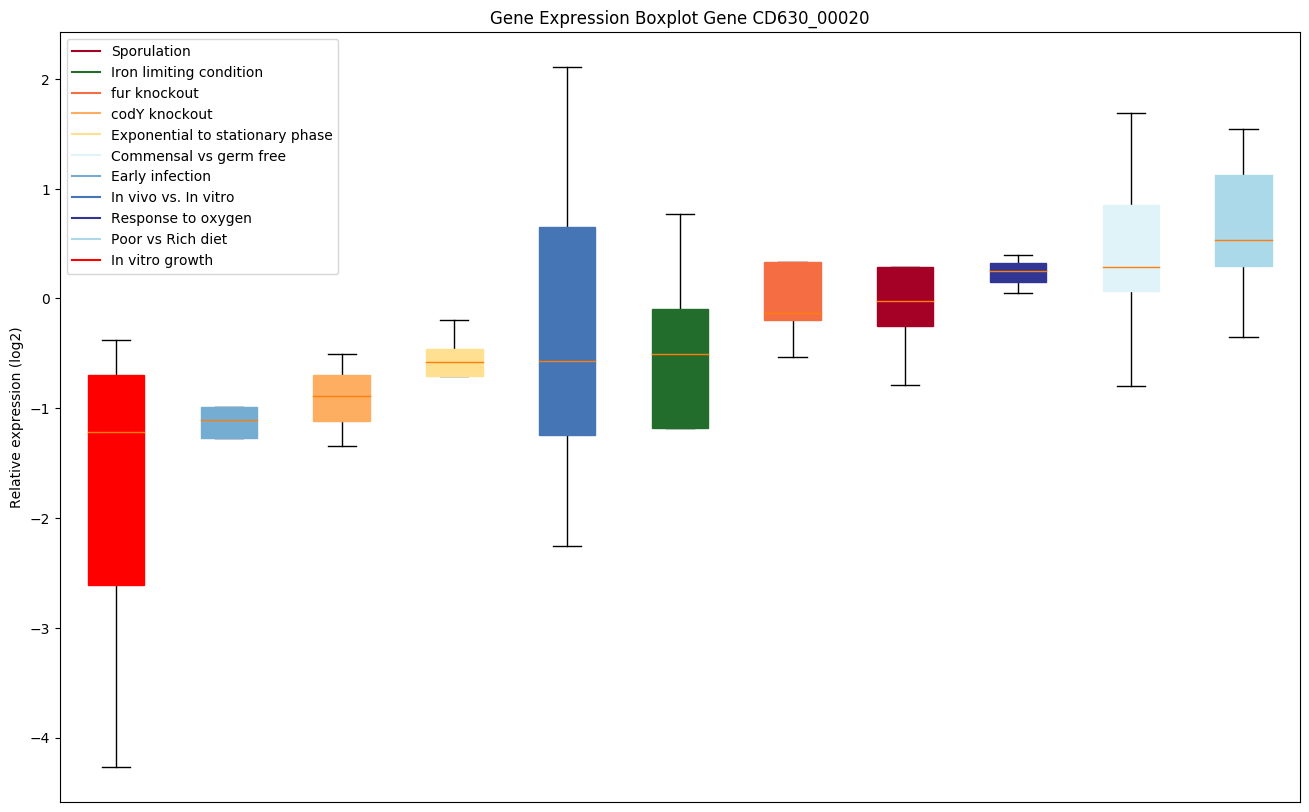 |
Bookmark | |||
| CD630_00030 | Putative RNA-binding mediating protein |
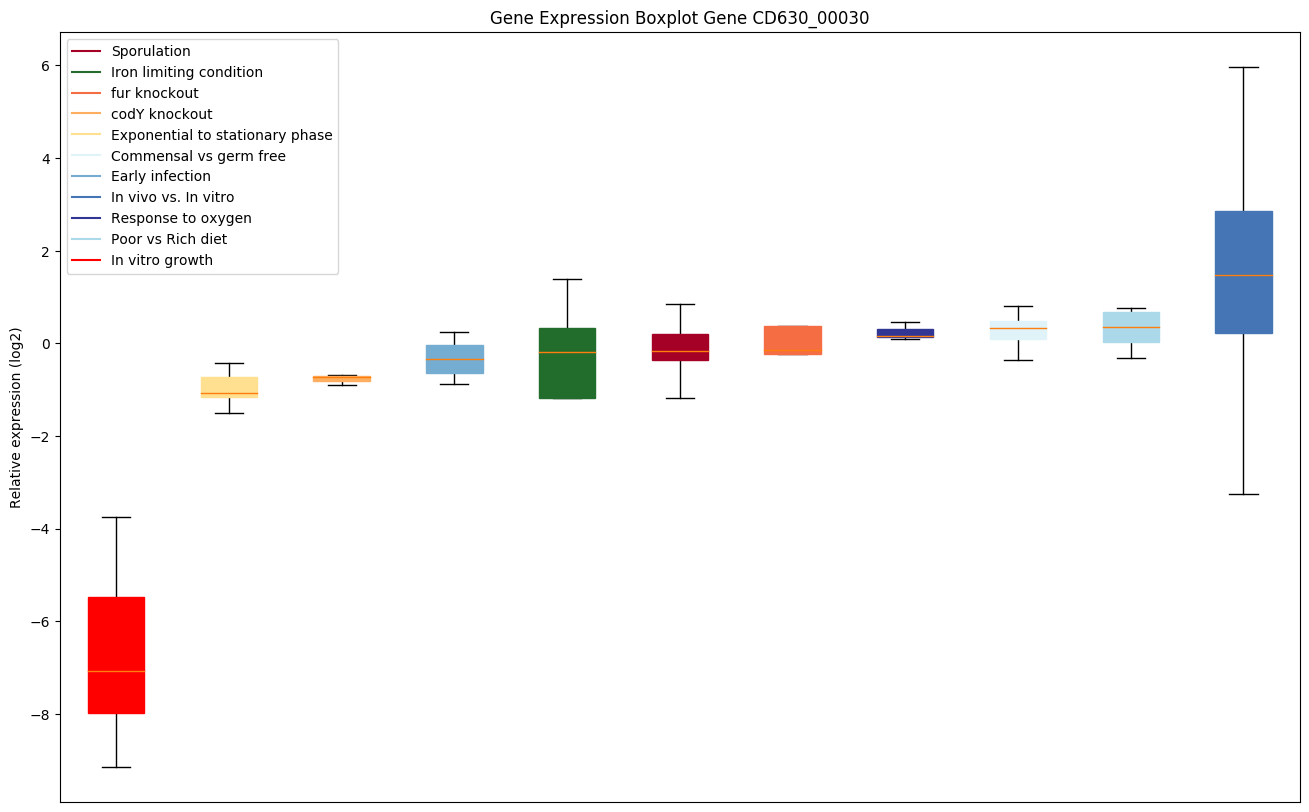 |
Bookmark | ||||
| CD630_00040 | recF | DNA replication and repair protein RecF |
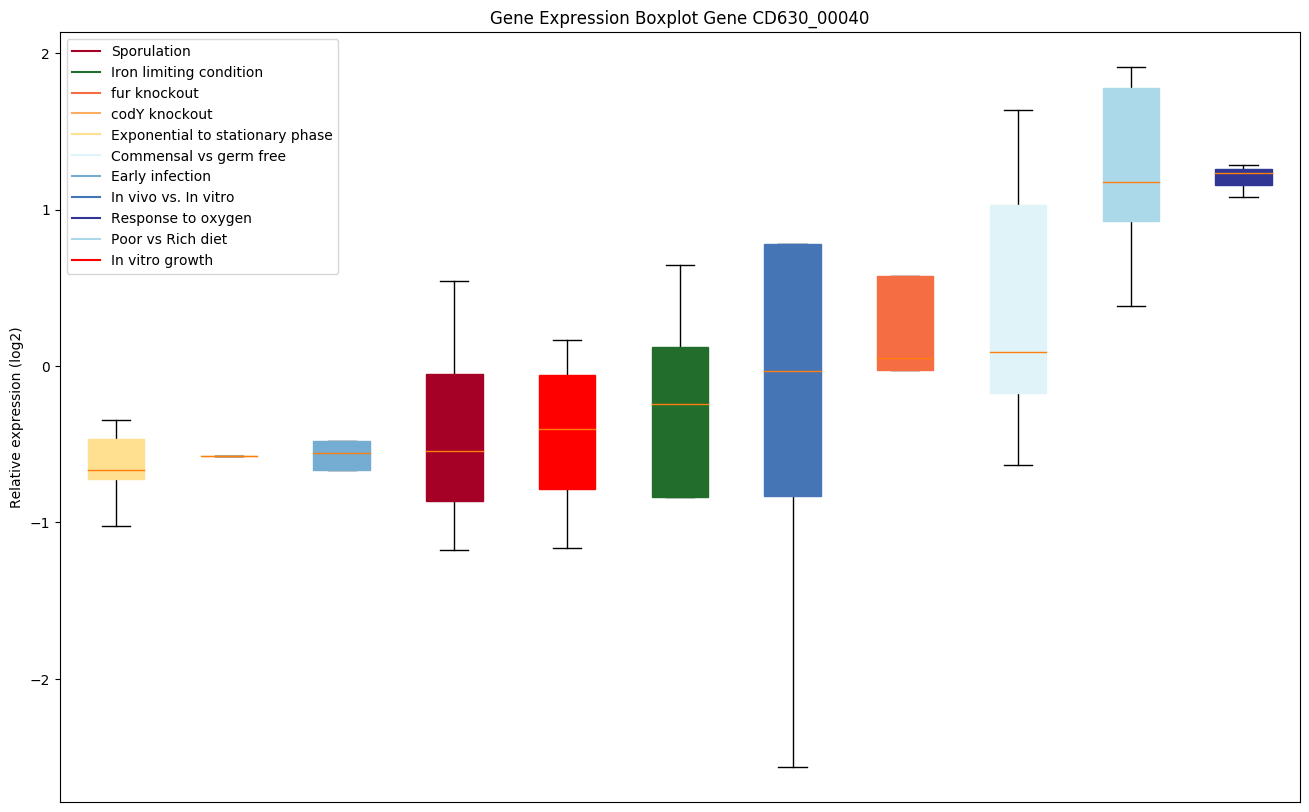 |
Bookmark | |||
| CD630_00050 | gyrB | DNA gyrase subunit B |
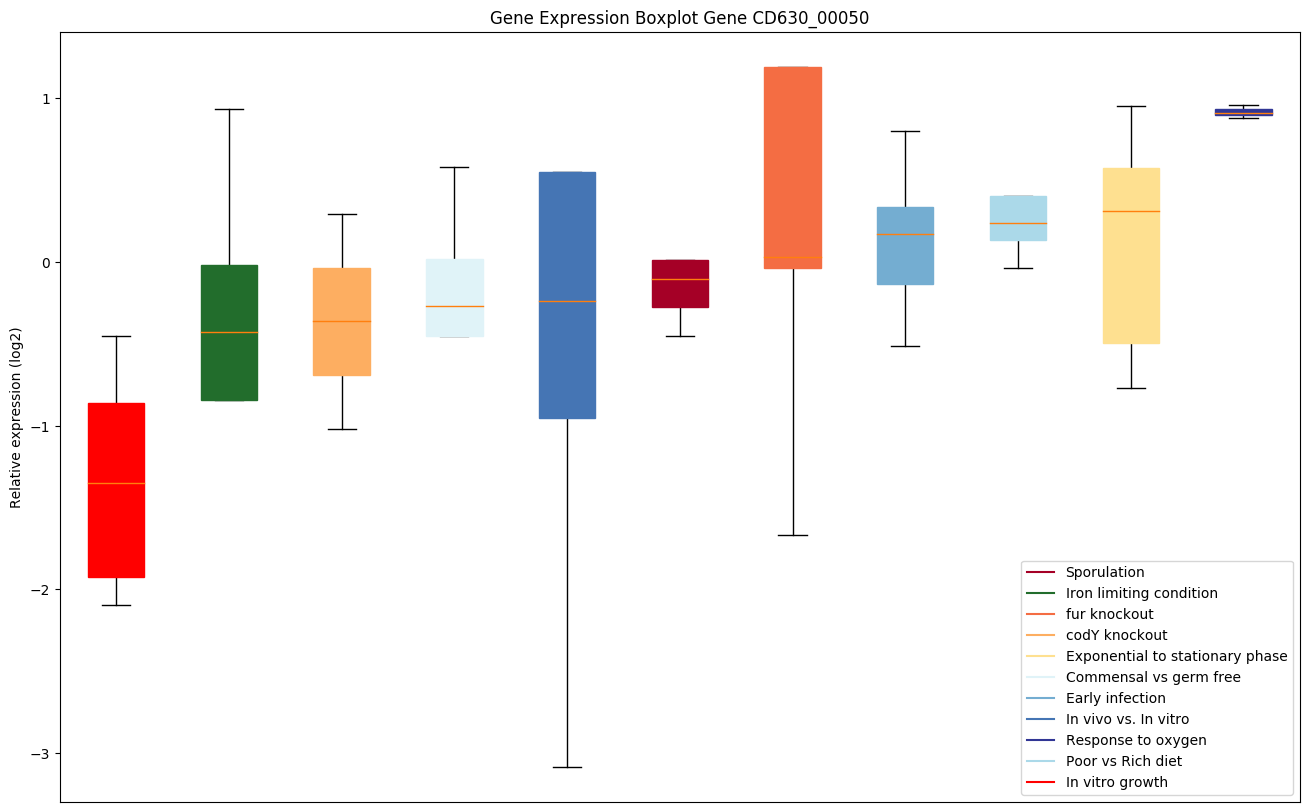 |
Bookmark | |||
| CD630_00060 | gyrA | DNA gyrase subunit A |
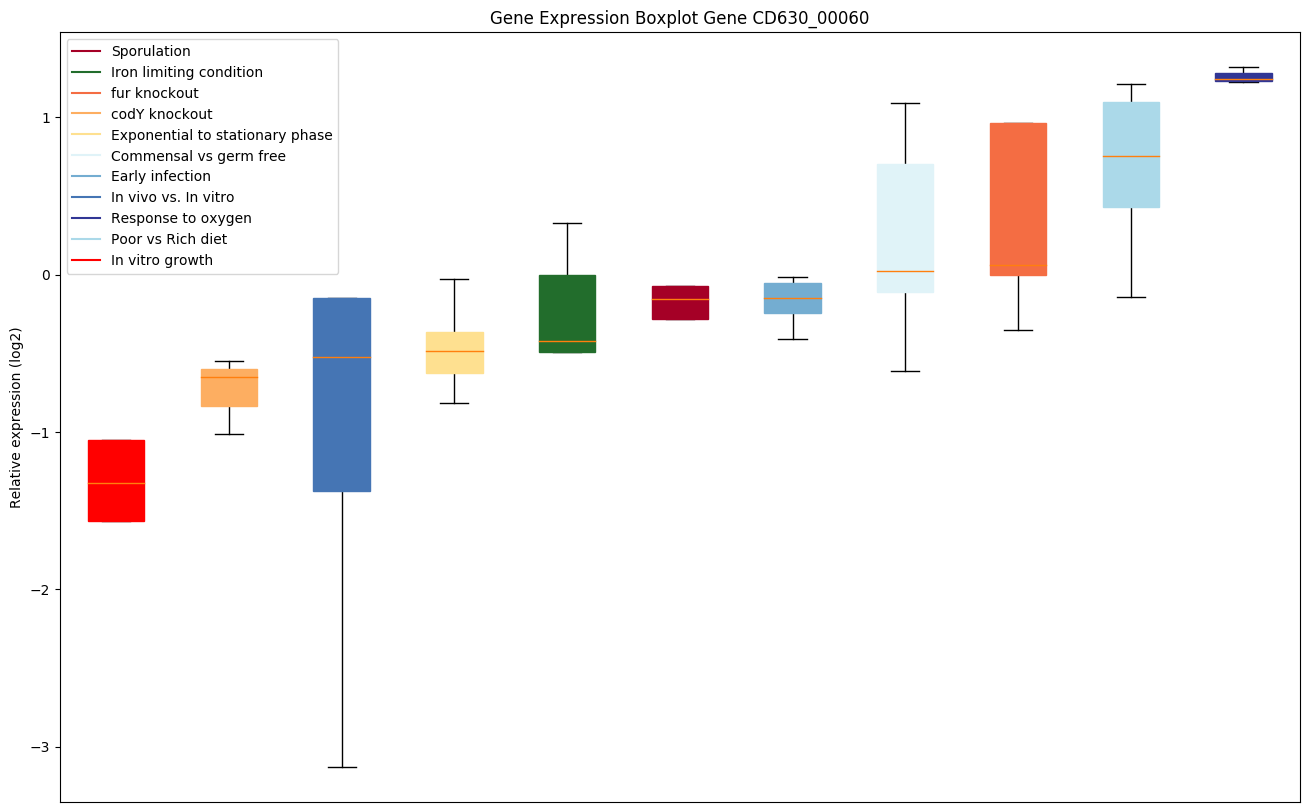 |
Bookmark | |||
| CD630_00070 | Conserved hypothetical protein |
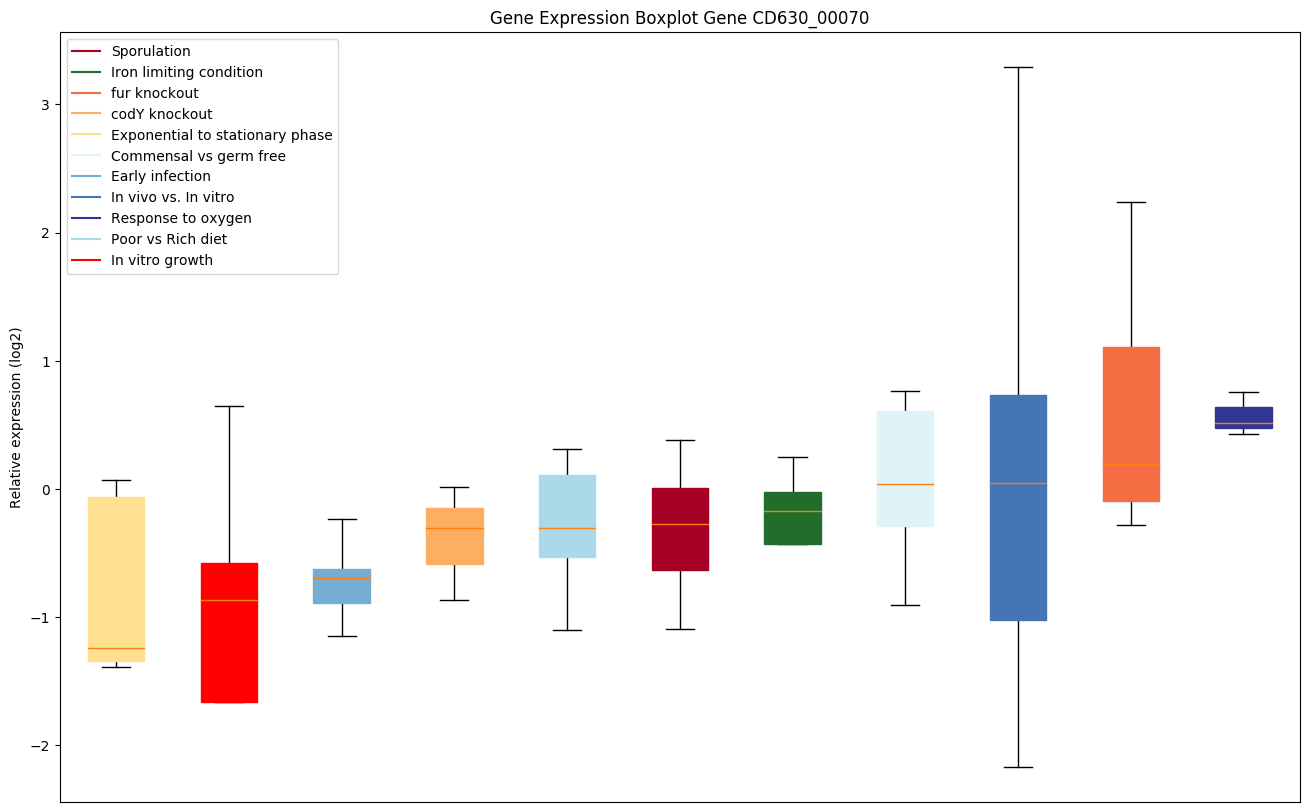 |
Bookmark | ||||
| CD630_00080 | Conserved hypothetical protein |
 |
Bookmark | ||||
| CD630_00090 | rsbV | Anti-anti-sigma factor |
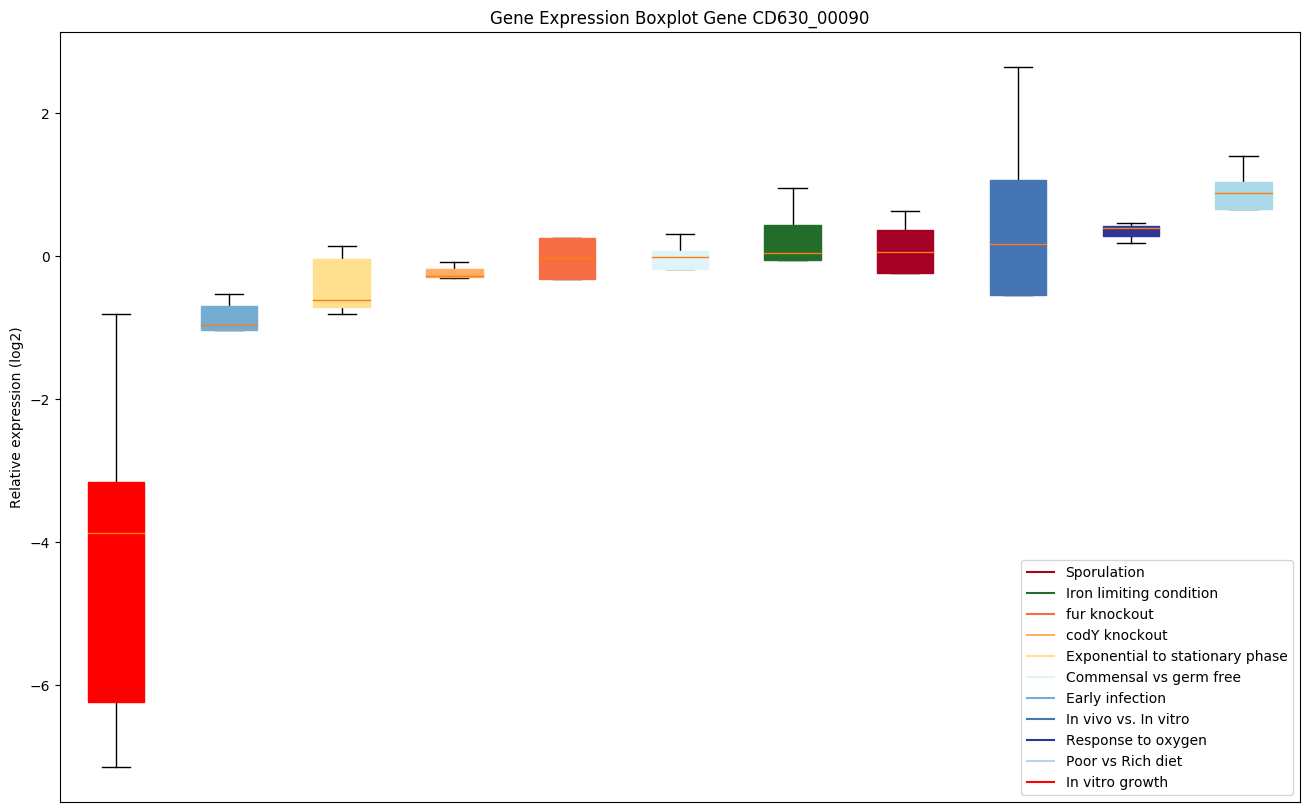 |
Bookmark | |||
| CD630_00100 | rsbW | Serine-protein kinase RsbW |
 |
Bookmark |
