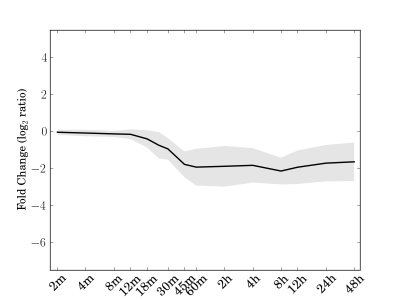
Module 23 Residual: 0.10
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0023 | v02 | 0.10 | -15.50 |
Displaying 1 - 26 of 26
| Cre01.g045902.t1.1 ATP binding | Cre03.g154600.t1.2 thylakoid lumen 15.0 kDa protein |
| Cre03.g156050.t1.2 ribosome recycling factor, chloroplast precursor | Cre03.g159500.t2.1 Pyridoxal-dependent decarboxylase family protein |
| Cre03.g183400.t1.1 | Cre05.g240850.t1.2 thiaminC |
| Cre06.g249750.t1.1 | Cre06.g296600.t1.2 lumazine-binding family protein |
| Cre06.g298350.t1.2 ARM repeat superfamily protein | Cre07.g334600.t1.2 Predicted protein |
| Cre08.g364450.t1.2 Acyl-CoA N-acyltransferases (NAT) superfamily protein | Cre10.g421600.t1.2 threonyl-tRNA synthetase, putative / threonine--tRNA ligase, putative |
| Cre12.g517900.t1.1 Albino or Glassy Yellow 1 | Cre12.g528000.t1.2 |
| Cre13.g579582.t1.1 starch synthase 3 | Cre13.g584850.t1.1 glutathione S-transferase PHI 10 |
| Cre13.g603900.t1.2 tRNA synthetase beta subunit family protein | Cre15.g641200.t1.2 Mitochondrial substrate carrier family protein |
| Cre16.g673617.t1.1 Peptide chain release factor 1 | Cre16.g678851.t1.1 Photosystem II reaction center PsbP family protein |
| Cre16.g684750.t1.2 NAD(P)-linked oxidoreductase superfamily protein | Cre16.g693850.t1.1 |
| Cre17.g738050.t1.2 PLAC8 family protein | Cre17.g739426.t1.1 nuclear shuttle interacting |
| Cre17.g741450.t1.2 chaperonin 60 beta | Cre17.g746547.t1.1 Basic-leucine zipper (bZIP) transcription factor family protein |
|
e.value: 0.00000000034 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: gAGCTgcgGaAGgAGCAGcAG |
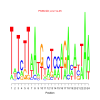
motif_0023_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.000021 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: TaTcTTaAtgtcTccaGAnantAA |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0006766 | GO:0006767 | GO:0009110 | GO:0042364 | GO:1901564 | GO:0004812 | vitamin metabolic process | water-soluble vitamin metabolic process | vitamin biosynthetic process | water-soluble vitamin biosynthetic proce... | organonitrogen compound metabolic proces... | aminoacyl-tRNA ligase activity |

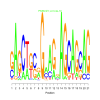
Comments