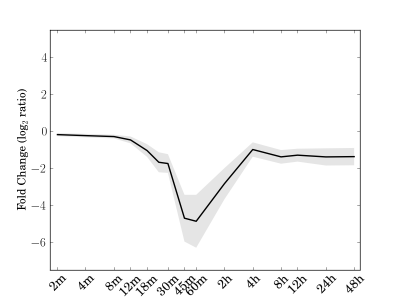
Module 126 Residual: 0.10
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0126 | v02 | 0.10 | -16.96 |
Displaying 1 - 17 of 17
| Cre01.g020950.t1.2 ribosome biogenesis regulatory protein (RRS1) family protein | Cre01.g044400.t1.2 |
| Cre01.g048950.t1.2 uridine 5\'-monophosphate synthase / UMP synthase (PYRE-F) (UMPS) | Cre02.g077300.t1.2 fibrillarin 2 |
| Cre03.g152950.t1.1 Pseudouridine synthase family protein | Cre03.g172950.t1.2 homologue of NAP57 |
| Cre06.g275100.t1.2 nucleolin like 2 | Cre07.g351750.t1.2 rRNA processing protein-related |
| Cre09.g392350.t2.1 glycine-rich RNA-binding protein 2 | Cre09.g416800.t1.2 |
| Cre10.g428750.t1.1 RNA binding;RNA binding | Cre10.g441400.t1.2 NOP56-like pre RNA processing ribonucleoprotein |
| Cre12.g483700.t1.2 organellar single-stranded DNA binding protein 3 | Cre12.g502300.t1.2 |
| Cre13.g562450.t1.2 ARM repeat superfamily protein | Cre16.g690500.t1.2 Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein |
| Cre16.g694202.t1.1 ubiquitin-specific protease 17 |
|
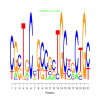
e.value: 0.001 Motif Bicluster: Width: 21 Number of Sites: 1 Consensus: CGcTgCcGccgcTGCtgCTGc |
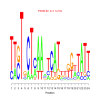
motif_0126_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 11000 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: TTgTgCTcAAatCATttggaTATT |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0022613 | GO:0042254 | GO:0006206 | GO:0006207 | GO:0019856 | GO:0004590 | ribonucleoprotein complex biogenesis | ribosome biogenesis | pyrimidine nucleobase metabolic process | 'de novo' pyrimidine nucleobase biosynth... | pyrimidine nucleobase biosynthetic proce... | orotidine-5'-phosphate decarboxylase act... |

Comments