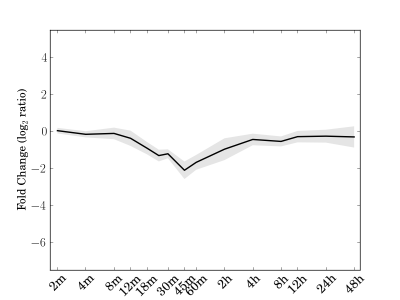
Module 188 Residual: 0.25
| Title | Model version | Residual | Score |
|---|---|---|---|
| bicluster_0188 | v02 | 0.25 | -12.40 |
Displaying 1 - 19 of 19
| Cre01.g027150.t1.1 DEAD/DEAH box helicase, putative | Cre01.g041400.t1.1 |
| Cre01.g054000.t1.2 D111/G-patch domain-containing protein | Cre03.g173000.t1.1 |
| Cre03.g178050.t1.1 Transducin/WD40 repeat-like superfamily protein | Cre04.g228150.t1.2 3-ketoacyl-CoA synthase 10 |
| Cre05.g238364.t1.1 | Cre06.g278171.t1.1 Protein of unknown function (DUF506) |
| Cre07.g312750.t1.2 Nucleic acid-binding, OB-fold-like protein | Cre07.g319900.t1.1 PNAS-3 related |
| Cre09.g387060.t1.1 organic cation/carnitine transporter1 | Cre09.g387726.t1.1 aspartate aminotransferase 3 |
| Cre09.g395600.t1.1 GTP1/OBG family protein | Cre09.g408500.t1.1 diphthamide synthesis DPH2 family protein |
| Cre10.g420000.t1.1 | Cre12.g497400.t1.2 RNA polymerase III RPC4 |
| Cre12.g543050.t1.2 | Cre16.g666334.t1.1 seed imbibition 2 |
| Cre17.g719900.t1.2 catalytics;carbohydrate kinases;phosphoglucan, water dikinases |
|
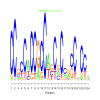
e.value: 0.00000041 Motif Bicluster: Width: 24 Number of Sites: 1 Consensus: CCgcCaCCGCCagCnCcgcCnCca |
motif_0188_2Submitted by Anonymous (not verified) on Wed, 05/20/2015 - 14:16e.value: 0.000000000039 Motif Bicluster: Width: 15 Number of Sites: 1 Consensus: CgGCAGCAGCAGCAG |
Displaying 1 - 1 of 1
| Interaction | Weight | |
|---|---|---|
Displaying 1 - 1 of 1
| GO Terms | Descriptions |
|---|---|
| GO:0006383 | transcription from RNA polymerase III pr... |

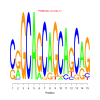
Comments