263246 PI-PLCc_eukaryotaThalassiosira pseudonana
| Chromosome | Product | Transcript Start | End | Strand | Short Name | |
|---|---|---|---|---|---|---|
| 263246 | chr_7 | PI-PLCc_eukaryota | 1486965 | 1488759 | + | PI-PLCc_eukaryota |
| NCBI ID | Ensembl Genomes exon ID |
|---|---|
| 7450698 | Thaps263246.5, Thaps263246.3, Thaps263246.2, Thaps263246.4, Thaps263246.7, Thaps263246.6, Thaps263246.1 |
| Expression Profile | Conditional Changes | Cluster Dendrogram | Discovered Potential cis-Regulatory Motifs |
|---|---|---|---|
Thaps_hclust_0391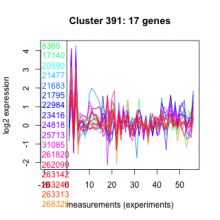 |
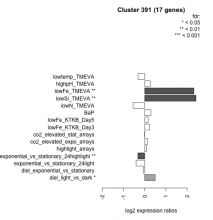 |
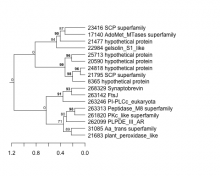 |
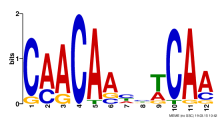  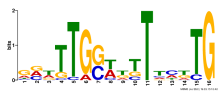 |
| T. pseudonana | P. tricornutum | P. tricornutum DiatomCyc | F. cylindrus | Pseudo-nitzschia multiseries | E. huxleyi | C. reinhardtii | A. thaliana | P. sojae |
|---|---|---|---|---|---|---|---|---|
| Not available | PHATRDRAFT_42683 | PHATRDRAFT_42683 | 193320 | 233390 | 248696 | Cre06.g270200.t1.1 | AT5G58690.1 | Not available |

Add comment