Details:
The cellular metabolic process in which a protein is formed, using the sequence of a mature mRNA molecule to specify the sequence of amino acids in a polypeptide chain. Translation is mediated by the ribosome, and begins with the formation of a ternary complex between aminoacylated initiator methionine tRNA, GTP, and initiation factor 2, which subsequently associates with the small subunit of the ribosome and an mRNA. Translation ends with the release of a polypeptide chain from the ribosome.


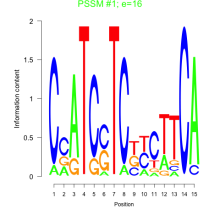
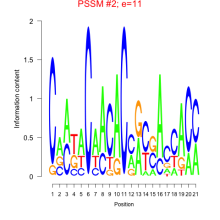

Add comment