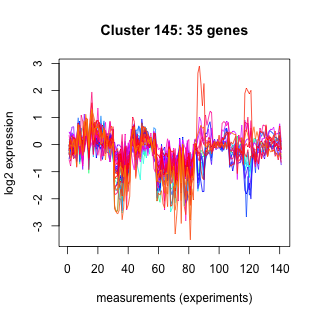
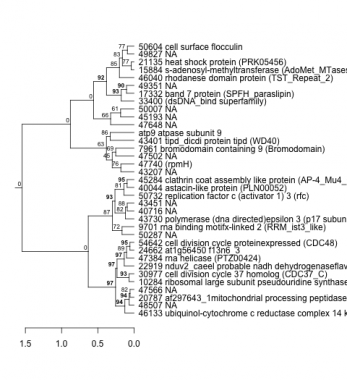
Phatr_hclust_0145 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 0.00296369 | 1 | Phatr_hclust_0145 |
| GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 0.00886694 | 1 | Phatr_hclust_0145 |
| GO:0006508 | proteolysis and peptidolysis | 0.0253471 | 1 | Phatr_hclust_0145 |
| GO:0006260 | DNA replication | 0.0744898 | 1 | Phatr_hclust_0145 |
| GO:0000074 | regulation of cell cycle | 0.0963646 | 1 | Phatr_hclust_0145 |
| GO:0006886 | intracellular protein transport | 0.112459 | 1 | Phatr_hclust_0145 |
| GO:0006511 | ubiquitin-dependent protein catabolism | 0.125669 | 1 | Phatr_hclust_0145 |
| GO:0006396 | RNA processing | 0.1541 | 1 | Phatr_hclust_0145 |
| GO:0006412 | protein biosynthesis | 0.385887 | 1 | Phatr_hclust_0145 |
| GO:0006118 | electron transport | 0.503223 | 1 | Phatr_hclust_0145 |
|
PHATRDRAFT_46133 : ubiquinol-cytochrome c reductase complex 14 kda (UCR_14kD superfamily) |
PHATRDRAFT_33400 : (dsDNA_bind superfamily) |
||
|
PHATRDRAFT_9701 : rna binding motifx-linked 2 (RRM_ist3_like) |
PHATRDRAFT_47740 : (rpmH) |
PHATRDRAFT_17332 : band 7 protein (SPFH_paraslipin) |
|
|
PHATRDRAFT_20787 : af297643_1mitochondrial processing peptidase beta subunit (PqqL) |
PHATRDRAFT_43730 : polymerase (dna directed)epsilon 3 (p17 subunit) (H4 superfamily) |
||
|
PHATRDRAFT_10284 : ribosomal large subunit pseudouridine synthase a (RluA) |
PHATRDRAFT_7961 : bromodomain containing 9 (Bromodomain) |
PHATRDRAFT_46040 : rhodanese domain protein (TST_Repeat_2) |
|
|
PHATRDRAFT_30977 : cell division cycle 37 homolog (CDC37_C) |
PHATRDRAFT_43401 : tipd_dicdi protein tipd (WD40) |
PHATRDRAFT_15884 : s-adenosyl-methyltransferase (AdoMet_MTases superfamily) |
|
|
PHATRDRAFT_22919 : nduv2_caeel probable nadh dehydrogenaseflavoproteinmitochondrial precursor (2Fe-2S_thioredx) |
PHATRDRAFT_50732 : replication factor c (activator 1) 3 (rfc) |
PHATRDRAFT_21135 : heat shock protein (PRK05456) |
|
|
PHATRDRAFT_47384 : rna helicase (PTZ00424) |
PHATRDRAFT_40044 : astacin-like protein (PLN00052) |
||
|
PHATRDRAFT_54642 : cell division cycle proteinexpressed (CDC48) |
PHATRDRAFT_45284 : clathrin coat assembly like protein (AP-4_Mu4_Cterm) |
PHATRDRAFT_50604 : cell surface flocculin |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | -0.812 | 0.000505051 |
| Copper_SH | Copper_SH | 0.191 | 0.0261261 |
| Green_vs_Red_24h | Green_vs_Red_24h | 0.171 | 0.0878505 |
| Silver_SH | Silver_SH | 0.052 | 0.522816 |
| Cadmium_1.2mg | Cadmium_1.2mg | 0.072 | 0.231915 |
| BlueLight_24h | BlueLight_24h | -0.326 | 0.00128205 |
| GreenLight_0.5h | GreenLight_0.5h | -0.995 | 0.000458716 |
| RedLight_6h | RedLight_6h | -0.863 | 0.000694444 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 1 |
| GreenLight_24h | GreenLight_24h | -0.433 | 0.000961538 |
| light_6hr | light_6hr | -0.166 | 0.402522 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | -0.037 | 0.862192 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | 0.092 | 0.255556 |
| Ammonia_SH | Ammonia_SH | -0.281 | 0.207328 |
| Simazine_SH | Simazine_SH | -0.010 | 0.923829 |
| Oil_SH | Oil_SH | -0.223 | 0.0141304 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | 0.055 | 0.555621 |
| Cadmium_SH | Cadmium_SH | -0.019 | 0.847374 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | 0.278 | 0.0345588 |
| highlight_0to6h | highlight_0to6h | 0.206 | 0.0424603 |
| BlueLight_0.5h | BlueLight_0.5h | -1.063 | 0.00338346 |
| Dispersant_SH | Dispersant_SH | -0.165 | 0.0669082 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | 0.098 | 0.328239 |
| light_15.5hr | light_15.5hr | 0.177 | 0.404899 |
| light_10.5hr | light_10.5hr | -0.123 | 0.603867 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.245 | 0.0107143 |
| Si_free | Si_free | -0.077 | 0.457971 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | -0.932 | 0.00206897 |
| Mixture_SH | Mixture_SH | -0.263 | 0.274309 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | -0.113 | 0.54375 |
| light_16hr_dark_30min | light_16hr_dark_30min | 0.214 | 0.293542 |
| BlueLight_6h | BlueLight_6h | -0.976 | 0.000581395 |
| highlight_12to48h | highlight_12to48h | 0.352 | 0.0138158 |
| RedLight_24h | RedLight_24h | -0.604 | 0.000943396 |
| Dark_treated | Dark_treated | -0.943 | 0.031746 |
| GreenLight_6h | GreenLight_6h | -1.068 | 0.000609756 |
| Cadmium_0.12mg | Cadmium_0.12mg | 0.057 | 0.065534 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | 0.107 | 0.0892473 |
| RedLight_0.5h | RedLight_0.5h | -1.120 | 0.000555556 |
| Green_vs_Red_6h | Green_vs_Red_6h | -0.205 | 0.183835 |
| Re-illuminated_24h | Re-illuminated_24h | -0.414 | 0.000657895 |