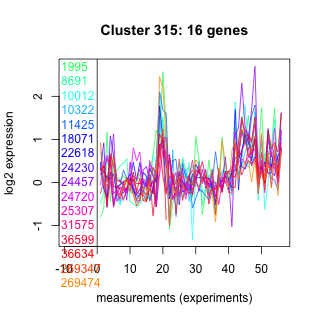
Thaps_hclust_0315 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006118 | electron transport | 0.00749022 | 1 | Thaps_hclust_0315 |
| GO:0051336 | regulation of hydrolase activity | 0.0115273 | 1 | Thaps_hclust_0315 |
| GO:0006520 | amino acid metabolism | 0.0342335 | 1 | Thaps_hclust_0315 |
| GO:0006281 | DNA repair | 0.073647 | 1 | Thaps_hclust_0315 |
| GO:0006508 | proteolysis and peptidolysis | 0.38827 | 1 | Thaps_hclust_0315 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.40843 | 1 | Thaps_hclust_0315 |
|
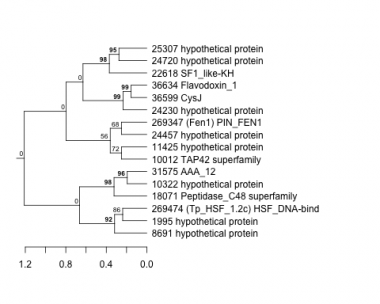
1995 : hypothetical protein |
11425 : hypothetical protein |
24457 : hypothetical protein |
36599 : CysJ |
|
8691 : hypothetical protein |
18071 : Peptidase_C48 superfamily |
24720 : hypothetical protein |
36634 : Flavodoxin_1 |
|
10012 : TAP42 superfamily |
22618 : SF1_like-KH |
25307 : hypothetical protein |
269347 : (Fen1) PIN_FEN1 |
|
10322 : hypothetical protein |
24230 : hypothetical protein |
31575 : AAA_12 |
269474 : (Tp_HSF_1.2c) HSF_DNA-bind |
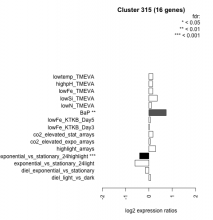
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | 0.077 | 0.807 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.014 | 0.968 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.061 | 0.814 |
| BaP | BaP | 0.741 | 0.00103 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.397 | 0.000926 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.177 | 0.499 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.174 | 0.565 |
| highpH_TMEVA | highpH_TMEVA | 0.160 | 0.381 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.056 | 0.903 |
| lowFe_TMEVA | lowFe_TMEVA | 0.171 | 0.624 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | -0.598 | 0.0801 |
| lowN_TMEVA | lowN_TMEVA | 0.113 | 0.784 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | -0.150 | 0.495 |
| lowSi_TMEVA | lowSi_TMEVA | 0.359 | 0.613 |
| highlight_arrays | highlight_arrays | 0.286 | 0.0981 |