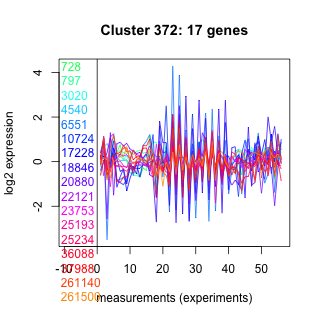
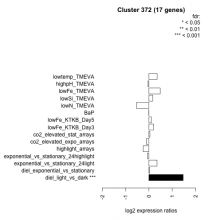
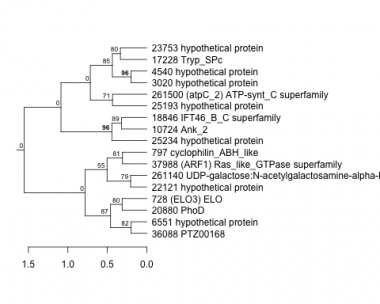
Thaps_hclust_0372 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015031 | protein transport | 0.068993 | 1 | Thaps_hclust_0372 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.073647 | 1 | Thaps_hclust_0372 |
| GO:0006886 | intracellular protein transport | 0.0859585 | 1 | Thaps_hclust_0372 |
| GO:0006457 | protein folding | 0.195964 | 1 | Thaps_hclust_0372 |
| GO:0006810 | transport | 0.266912 | 1 | Thaps_hclust_0372 |
| GO:0006508 | proteolysis and peptidolysis | 0.38827 | 1 | Thaps_hclust_0372 |
| GO:0008152 | metabolism | 0.505245 | 1 | Thaps_hclust_0372 |
| GO:0007264 | small GTPase mediated signal transduction | 0.0549078 | 1 | Thaps_hclust_0372 |
|
728 : (ELO3) ELO |
10724 : Ank_2 |
22121 : hypothetical protein |
36088 : PTZ00168 |
|
797 : cyclophilin_ABH_like |
17228 : Tryp_SPc |
23753 : hypothetical protein |
37988 : (ARF1) Ras_like_GTPase superfamily |
|
3020 : hypothetical protein |
18846 : IFT46_B_C superfamily |
25193 : hypothetical protein |
261140 : UDP-galactose:N-acetylgalactosamine-alpha-R beta 1,3-galactosyltransferase |
|
4540 : hypothetical protein |
20880 : PhoD |
25234 : hypothetical protein |
261500 : (atpC_2) ATP-synt_C superfamily |
|
6551 : hypothetical protein |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | 1.470 | 0.000485 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.202 | 0.464 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.106 | 0.65 |
| BaP | BaP | -0.005 | 0.994 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.055 | 0.676 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.045 | 0.884 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.354 | 0.206 |
| highpH_TMEVA | highpH_TMEVA | -0.038 | 0.867 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | -0.105 | 0.695 |
| lowFe_TMEVA | lowFe_TMEVA | 0.487 | 0.0729 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.349 | 0.318 |
| lowN_TMEVA | lowN_TMEVA | -0.530 | 0.122 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.037 | 0.884 |
| lowSi_TMEVA | lowSi_TMEVA | 0.163 | 0.939 |
| highlight_arrays | highlight_arrays | -0.246 | 0.151 |