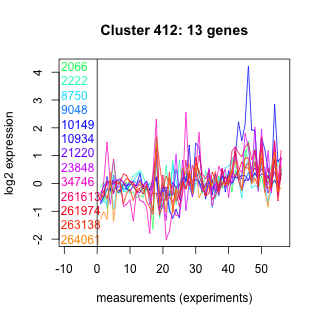
Thaps_hclust_0412 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006370 | mRNA capping | 0.00557274 | 1 | Thaps_hclust_0412 |
| GO:0000103 | sulfate assimilation | 0.00742418 | 1 | Thaps_hclust_0412 |
| GO:0030163 | protein catabolism | 0.0239497 | 1 | Thaps_hclust_0412 |
| GO:0006397 | mRNA processing | 0.0402303 | 1 | Thaps_hclust_0412 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0473882 | 1 | Thaps_hclust_0412 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.107561 | 1 | Thaps_hclust_0412 |
| GO:0006508 | proteolysis and peptidolysis | 0.424754 | 1 | Thaps_hclust_0412 |
| GO:0008152 | metabolism | 0.546944 | 1 | Thaps_hclust_0412 |
|
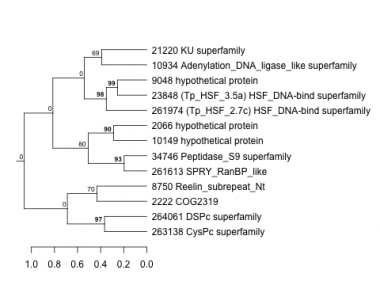
2066 : hypothetical protein |
10149 : hypothetical protein |
23848 : (Tp_HSF_3.5a) HSF_DNA-bind superfamily |
261974 : (Tp_HSF_2.7c) HSF_DNA-bind superfamily |
|
2222 : COG2319 |
10934 : Adenylation_DNA_ligase_like superfamily |
34746 : Peptidase_S9 superfamily |
263138 : CysPc superfamily |
|
8750 : Reelin_subrepeat_Nt |
21220 : KU superfamily |
261613 : SPRY_RanBP_like |
264061 : DSPc superfamily |
|
9048 : hypothetical protein |
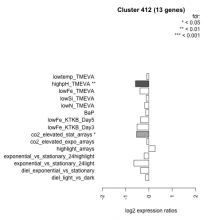
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | -0.135 | 0.67 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.505 | 0.0815 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.359 | 0.157 |
| BaP | BaP | -0.132 | 0.656 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.204 | 0.122 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.531 | 0.0378 |
| lowtemp_TMEVA | lowtemp_TMEVA | -0.091 | 0.799 |
| highpH_TMEVA | highpH_TMEVA | -0.577 | 0.00357 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | -0.091 | 0.802 |
| lowFe_TMEVA | lowFe_TMEVA | -0.385 | 0.226 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | -0.642 | 0.0894 |
| lowN_TMEVA | lowN_TMEVA | -0.181 | 0.67 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | -0.356 | 0.109 |
| lowSi_TMEVA | lowSi_TMEVA | -0.118 | 0.976 |
| highlight_arrays | highlight_arrays | 0.264 | 0.173 |