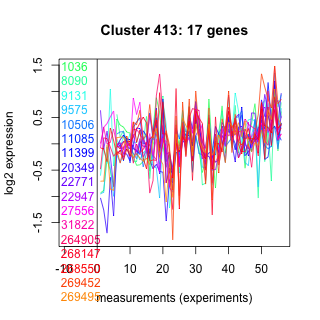
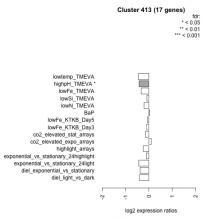
Thaps_hclust_0413 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006865 | amino acid transport | 0.0470024 | 1 | Thaps_hclust_0413 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.073647 | 1 | Thaps_hclust_0413 |
| GO:0006118 | electron transport | 0.364179 | 1 | Thaps_hclust_0413 |
| GO:0008152 | metabolism | 0.505245 | 1 | Thaps_hclust_0413 |
| GO:0009401 | phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.0294066 | 1 | Thaps_hclust_0413 |
| GO:0016567 | protein ubiquitination | 0.123514 | 1 | Thaps_hclust_0413 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.40843 | 1 | Thaps_hclust_0413 |
| GO:0006512 | ubiquitin cycle | 0.0580538 | 1 | Thaps_hclust_0413 |
|
1036 : hypothetical protein |
11085 : hypothetical protein |
22947 : zf-UBR |
268147 : Trp_Tyr_perm superfamily |
|
8090 : hypothetical protein |
11399 : hypothetical protein |
27556 : HSH155 |
268550 : (ALD1) ALDH-SF superfamily |
|
9131 : SCP superfamily |
20349 : GH38-57_N_LamB_YdjC_SF superfamily |
31822 : WD40 |
269452 : hypothetical protein |
|
9575 : hypothetical protein |
22771 : hypothetical protein |
264905 : FeFe_hydrog_A |
269495 : hypothetical protein |
|
10506 : hypothetical protein |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | -0.407 | 0.0558 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.098 | 0.763 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.058 | 0.82 |
| BaP | BaP | 0.038 | 0.908 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.114 | 0.349 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.150 | 0.574 |
| lowtemp_TMEVA | lowtemp_TMEVA | -0.457 | 0.102 |
| highpH_TMEVA | highpH_TMEVA | -0.422 | 0.0185 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.104 | 0.705 |
| lowFe_TMEVA | lowFe_TMEVA | -0.330 | 0.26 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | -0.433 | 0.198 |
| lowN_TMEVA | lowN_TMEVA | -0.206 | 0.594 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | -0.374 | 0.0509 |
| lowSi_TMEVA | lowSi_TMEVA | -0.088 | 0.997 |
| highlight_arrays | highlight_arrays | -0.238 | 0.165 |