Phatr_bicluster_0006 Residual: 0.56
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0006 | 0.56 | Phaeodactylum tricornutum |
Displaying 1 - 70 of 70
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_14945 v-typec subunit family protein (ATP-synt_C superfamily)
PHATRDRAFT_15692 golgi membrane protein sb140 isoform 1 (YIP1 superfamily)
PHATRDRAFT_16442 ormdl family protein (ORMDL superfamily)
PHATRDRAFT_16571 vacuolar type h+-atpase proteolipid subunit (ATP-synt_C superfamily)
PHATRDRAFT_16719 phosphoglucomutase 2 (PGM2)
PHATRDRAFT_16854 hcf101 (high-chlorophyll-fluorescence 101) atp binding (MRP-like)
PHATRDRAFT_19329 potential glutaredoxin (GRX_GRXh_1_2_like)
PHATRDRAFT_21207 (PLN02820)
PHATRDRAFT_22398 abc1 family protein (AarF)
PHATRDRAFT_22677 lactation elevated 1 (AAA superfamily)
PHATRDRAFT_2966 degenerative spermatocyte homolog 1 (PLN02579)
PHATRDRAFT_34419 aminotransferase class v (PLN02651)
PHATRDRAFT_38754 loc496288 protein
PHATRDRAFT_38948 (DUF92 superfamily)
PHATRDRAFT_40174 subfamilymember 5 (DnaJ)
PHATRDRAFT_40830 (LRR_RI superfamily)
PHATRDRAFT_40956 gtp-binding nuclear protein ran (PTZ00132)
PHATRDRAFT_40988 cgi-122 protein (GINS_A_psf2)
PHATRDRAFT_41125 chaperonin containingsubunit 8isoform 5 (TCP1_theta)
PHATRDRAFT_41541 enoyl-hydratase isomerase (PRK07509)
PHATRDRAFT_42828 glutathione s-transferase domain protein (ECM4)
PHATRDRAFT_44152 rna recognition(or rnp) domain containing protein (RRM_SF superfamily)
PHATRDRAFT_4593 major facilitator family transporter (MFS_1)
PHATRDRAFT_46447 alpha beta hydrolase fold (Abhydrolase_6)
PHATRDRAFT_47042 hcf101 (high-chlorophyll-fluorescence 101) atp binding (MRP-like)
PHATRDRAFT_47432 kda class i heat shock protein (ACD_sHsps-like)
PHATRDRAFT_48041 riboflavin-specific deaminase-like protein (RibD)
PHATRDRAFT_48208 metacaspase (Peptidase_C14)
PHATRDRAFT_48660 (PDZ superfamily)
PHATRDRAFT_48855 (Cyt_b561_FRRS1_like)
PHATRDRAFT_49564 nucleolar protein 9 (COG1341)
PHATRDRAFT_49750 ankyrin unc44 (ANK)
PHATRDRAFT_49932 helicase domain protein (HA)
PHATRDRAFT_50047 (CIA30 superfamily)
PHATRDRAFT_50216 protein phosphataseregulatory subunit 1
PHATRDRAFT_5043 serine incorporatorisoform cra_a (Serinc superfamily)
PHATRDRAFT_50562 astacin-like protein (PLN00052)
PHATRDRAFT_50572 ubiquitin-activating enzyme e1 1 (E1_enzyme_family superfamily)
PHATRDRAFT_5537 er lumen protein retaining receptor (ER_lumen_recept)
PHATRDRAFT_5629 membrane protein (Cyt_b561_FRRS1_like)
PHATRDRAFT_8772 principal rna polymerase sigma factor (Sigma70_r2)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015031 | protein transport | 0.0189497 | 1 | Phatr_bicluster_0006 |
| GO:0015986 | ATP synthesis coupled proton transport | 0.0264586 | 1 | Phatr_bicluster_0006 |
| GO:0009073 | aromatic amino acid family biosynthesis | 0.0341181 | 1 | Phatr_bicluster_0006 |
| GO:0045454 | cell redox homeostasis | 0.0408058 | 1 | Phatr_bicluster_0006 |
| GO:0009231 | riboflavin biosynthesis | 0.0408058 | 1 | Phatr_bicluster_0006 |
| GO:0044267 | cellular protein metabolism | 0.0671124 | 1 | Phatr_bicluster_0006 |
| GO:0006636 | fatty acid desaturation | 0.0671124 | 1 | Phatr_bicluster_0006 |
| GO:0006352 | transcription initiation | 0.0800035 | 1 | Phatr_bicluster_0006 |
| GO:0006725 | aromatic compound metabolism | 0.129871 | 1 | Phatr_bicluster_0006 |
| GO:0007264 | small GTPase mediated signal transduction | 0.135918 | 1 | Phatr_bicluster_0006 |

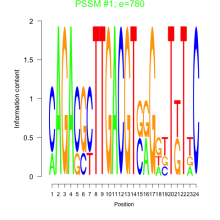

Comments