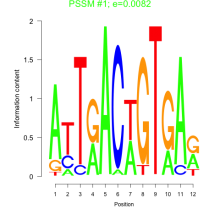
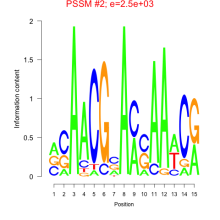
Phatr_bicluster_0034 Residual: 0.35
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0034 | 0.35 | Phaeodactylum tricornutum |
Displaying 1 - 38 of 38
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_10518 5-adenylylsulfate reductase (Thioredoxin_like superfamily)
PHATRDRAFT_13107 peptide methionine sulfoxide reductase (MsrA)
PHATRDRAFT_14478 adrenodoxin-type ferredoxin (fer2)
PHATRDRAFT_1884 leucine aminopeptidase (PRK00913)
PHATRDRAFT_23444 gtp-binding protein typa (TypA)
PHATRDRAFT_23717 chloroplast ferredoxin nadp(+) reductase (PLN03116)
PHATRDRAFT_23850 at1g34430 f7p12_2 (PRK11856)
PHATRDRAFT_27323 tyrosyl-trna synthetase (PRK05912)
PHATRDRAFT_34010 alanine aminotransferase (PTZ00377)
PHATRDRAFT_37835 (Dynamitin)
PHATRDRAFT_40467 m-like protein
PHATRDRAFT_43560 2-nitropropane dioxygenase npd (NPD_like)
PHATRDRAFT_44384 4-hydroxyphenylpyruvate dioxygenase (4HPPD)
PHATRDRAFT_45240 gcn5-related n-acetyltransferase
PHATRDRAFT_45840 c-type u1 snrnp
PHATRDRAFT_47395 thylakoid ascorbate peroxidase (ascorbate_peroxidase)
PHATRDRAFT_47449 lsu ribosomal protein l9p (rplI)
PHATRDRAFT_47505 (Exostosin superfamily)
PHATRDRAFT_48078 carnitine acetyl transferase (Carn_acyltransf superfamily)
PHATRDRAFT_48683 ankyrin unc44 (ANK)
PHATRDRAFT_49127 (RanBD_family)
PHATRDRAFT_49494 zinc finger (c3hc4-type ring finger) family protein (RING)
PHATRDRAFT_49965 cg9520-isoform b
PHATRDRAFT_50489 agc family protein kinase (S_TKc)
PHATRDRAFT_54738 triosephosphate isomerase (TIM)
PHATRDRAFT_8024 ribosomal protein l22 (Ribosomal_L22)
PHATRDRAFT_9400 peptidyl-prolyl cis-transfkbp-type (FkpA)
PHATRDRAFT_9701 rna binding motifx-linked 2 (RRM_ist3_like)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006437 | tyrosyl-tRNA aminoacylation | 0.00444554 | 1 | Phatr_bicluster_0034 |
| GO:0006118 | electron transport | 0.0162729 | 1 | Phatr_bicluster_0034 |
| GO:0006412 | protein biosynthesis | 0.032453 | 1 | Phatr_bicluster_0034 |
| GO:0006979 | response to oxidative stress | 0.101687 | 1 | Phatr_bicluster_0034 |
| GO:0019538 | protein metabolism | 0.148789 | 1 | Phatr_bicluster_0034 |
| GO:0009058 | biosynthesis | 0.182574 | 1 | Phatr_bicluster_0034 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.207972 | 1 | Phatr_bicluster_0034 |
| GO:0006464 | protein modification | 0.246382 | 1 | Phatr_bicluster_0034 |
| GO:0016567 | protein ubiquitination | 0.276493 | 1 | Phatr_bicluster_0034 |
| GO:0006457 | protein folding | 0.410758 | 1 | Phatr_bicluster_0034 |

Comments