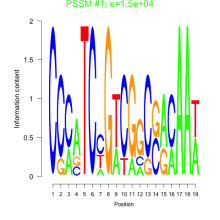
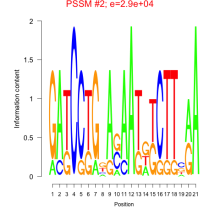
Phatr_bicluster_0051 Residual: 0.31
Phaeodactylum tricornutum
| Title | Residual | Model version | Organism |
|---|---|---|---|
| Phatr_bicluster_0051 | 0.31 | Phaeodactylum tricornutum |
Displaying 1 - 27 of 27
" class="views-fluidgrid-wrapper clear-block">
PHATRDRAFT_11916 1-acyl-sn-glycerol-3-phosphate acyltransferase (LPLAT_AGPAT-like)
PHATRDRAFT_13725 iaa amidohydrolase (M20_Acy1)
PHATRDRAFT_14552 trna-ia37 thiotransferase enzyme (PRK14333)
PHATRDRAFT_16803 (AarF)
PHATRDRAFT_16963 glutaminyl-trna synthetase (PRK05347)
PHATRDRAFT_30636 (DUF4336)
PHATRDRAFT_32923 inorganic pyrophosphatase (pyrophosphatase)
PHATRDRAFT_34146 60s ribosomal protein l66 (KOW_RPL6)
PHATRDRAFT_42636 transportin-serine arginine rich cg2848-pa (Xpo1 superfamily)
PHATRDRAFT_43194 ae family transporter: anion exchange (HCO3_cotransp superfamily)
PHATRDRAFT_43940 mitochondrial transcription termination factor family protein mterf family protein (mTERF superfamily)
PHATRDRAFT_45818 (MFS)
PHATRDRAFT_46330 (Mem_trans superfamily)
PHATRDRAFT_49533 plastidic atp adp transporter (TLC)
PHATRDRAFT_49843 mopfamily transporter (MATE_like superfamily)
PHATRDRAFT_54495 iron-responsive transporter-related (FPN1 superfamily)
PHATRDRAFT_55177 auman spermidine synthase (AdoMet_MTases)
PHATRDRAFT_573 inositol 5-phosphatase (EEP superfamily)
PHATRDRAFT_8112 cdp diglyceride synthetase cg7962-pa (CTP_transf_1 superfamily)
PHATRDRAFT_8926 aminoacyl-trna synthetase cofactor (tRNA_bindingDomain superfamily)
PHATRDRAFT_9860 phenylalanyl-trnaalpha subunit (pheS)
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0008654 | phospholipid biosynthesis | 0.00039425 | 0.902043 | Phatr_bicluster_0051 |
| GO:0006424 | glutamyl-tRNA aminoacylation | 0.00690418 | 1 | Phatr_bicluster_0051 |
| GO:0006432 | phenylalanyl-tRNA aminoacylation | 0.0103397 | 1 | Phatr_bicluster_0051 |
| GO:0006418 | tRNA aminoacylation for protein translation | 0.0133392 | 1 | Phatr_bicluster_0051 |
| GO:0006820 | anion transport | 0.0407661 | 1 | Phatr_bicluster_0051 |
| GO:0006412 | protein biosynthesis | 0.104611 | 1 | Phatr_bicluster_0051 |
| GO:0008152 | metabolism | 0.270353 | 1 | Phatr_bicluster_0051 |
| GO:0006468 | protein amino acid phosphorylation | 0.455951 | 1 | Phatr_bicluster_0051 |
| GO:0006810 | transport | 0.492171 | 1 | Phatr_bicluster_0051 |
| GO:0006508 | proteolysis and peptidolysis | 0.549807 | 1 | Phatr_bicluster_0051 |

Comments